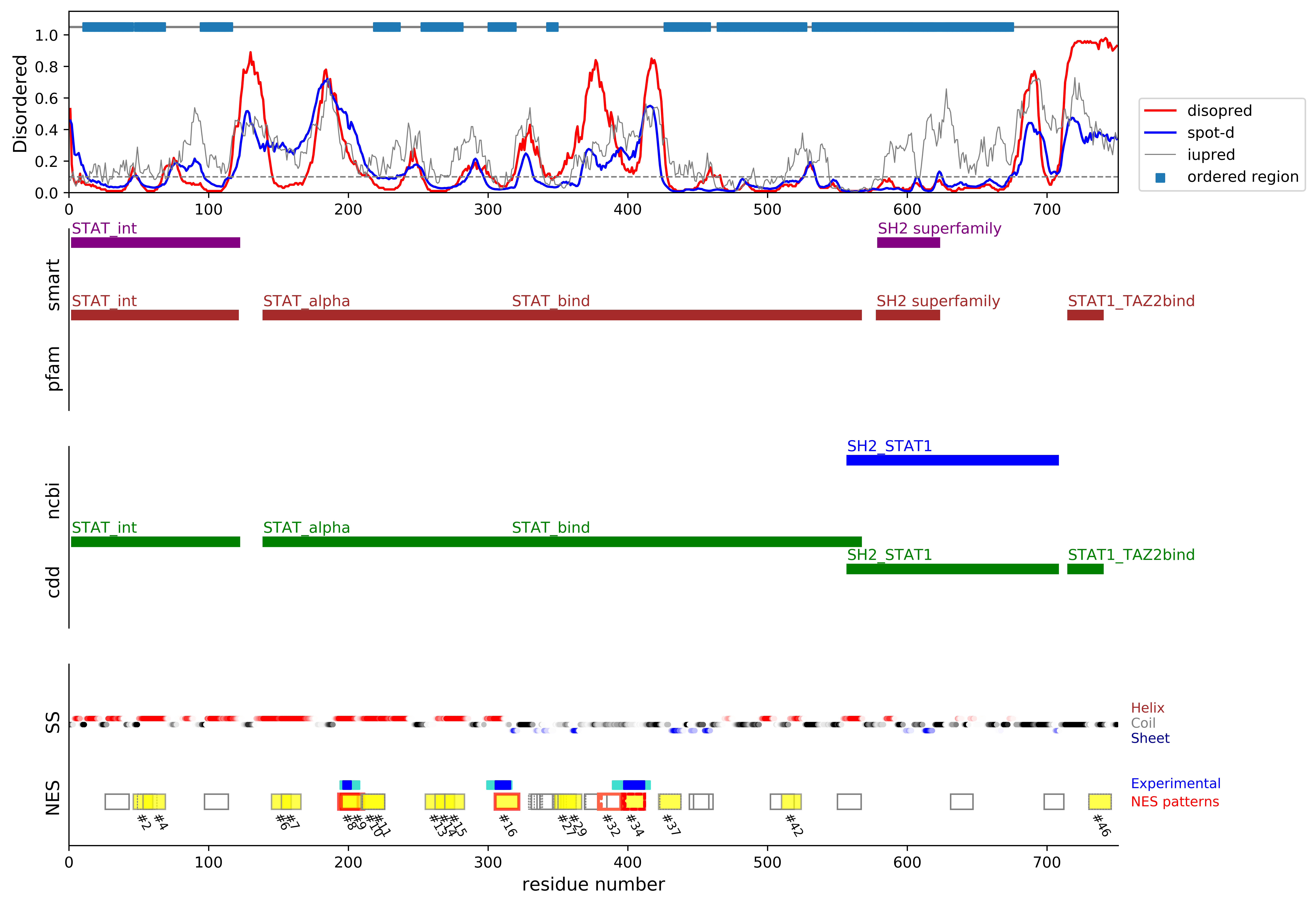

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P42224 | 26 | FPMEIRQYLAQWLEKQD | CCHHHHHHHHHHHHCCC | c1cR-4 | uniq | 0.016 | 0.04 | 0.132 | boundary | MID|STAT_int; | 0.0 |
| 2 | fp_D | P42224 | 46 | AANDVSFATIRFHD | CCCCHHHHHHHHHH | c2-4 | multi-selected | 0.059 | 0.065 | 0.158 | boundary | MID|STAT_int; | 0.0 |
| 3 | fp_D | P42224 | 49 | DVSFATIRFHDLLS | CHHHHHHHHHHHHH | c3-AT | multi | 0.033 | 0.049 | 0.142 | boundary | MID|STAT_int; | 0.0 |
| 4 | fp_D | P42224 | 53 | ATIRFHDLLSQLDDQY | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.038 | 0.039 | 0.129 | boundary | MID|STAT_int; | 0.0 |
| 5 | fp_D | P42224 | 97 | DPIQMSMIIYSCLKEER | CCHHHHHHHHHHHHHHH | c1cR-4 | uniq | 0.018 | 0.06 | 0.191 | boundary | boundary|STAT_int; | 0.0 |
| 6 | fp_D | P42224 | 145 | KVRNVKDKVMCIEH | HHHHHHHHHHHHHH | c3-4 | uniq | 0.053 | 0.287 | 0.289 | DISO | boundary|STAT_int; boundary|STAT_alpha; | 0.0 |
| 7 | fp_D | P42224 | 152 | KVMCIEHEIKSLED | HHHHHHHHHHHHHH | c3-4 | uniq | 0.046 | 0.277 | 0.238 | DISO | boundary|STAT_int; boundary|STAT_alpha; | 0.0 |
| 8 | cand_D | P42224 | 193 | KQEQLLLKKMYLML ++*++++++ | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.283 | 0.46 | 0.269 | boundary | MID|STAT_alpha; | 0.0 |
| 9 | cand_D | P42224 | 195 | EQLLLKKMYLMLDNKR ++*++++++ | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.222 | 0.406 | 0.228 | boundary | MID|STAT_alpha; | 0.0 |
| 10 | fp_D | P42224 | 209 | KRKEVVHKIIELLNVTE | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.075 | 0.142 | 0.172 | boundary | MID|STAT_alpha; | 0.0 |
| 11 | fp_D | P42224 | 210 | RKEVVHKIIELLNVTE | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.072 | 0.134 | 0.172 | boundary | MID|STAT_alpha; | 0.0 |
| 12 | fp_D | P42224 | 210 | RKEVVHKIIELLNVTE | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.072 | 0.134 | 0.172 | boundary | MID|STAT_alpha; | 0.0 |
| 13 | fp_D | P42224 | 255 | CLDQLQNWFTIVAE | CCCCCCCHHHHHHH | c3-4 | uniq | 0.017 | 0.046 | 0.114 | boundary | MID|STAT_alpha; | 0.0 |
| 14 | fp_D | P42224 | 262 | WFTIVAESLQQVRQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.016 | 0.031 | 0.113 | boundary | MID|STAT_alpha; | 0.0 |
| 15 | fp_D | P42224 | 269 | SLQQVRQQLKKLEE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.051 | 0.041 | 0.2 | boundary | MID|STAT_alpha; | 0.0 |
| 16 | cand_D | P42224 | 305 | TFSLFQQLIQSSFVVER +++**++**+ | HHHHHHHHHCCEEEEEE | c1c-5 | uniq | 0.078 | 0.03 | 0.122 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.125 |
| 17 | fp_beta_D | P42224 | 329 | PQRPLVLKTGVQFTVKL | CCCCEEEEECCEEEEEE | c1c-AT-4 | multi | 0.219 | 0.083 | 0.243 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.75 |
| 18 | fp_beta_D | P42224 | 330 | QRPLVLKTGVQFTVKL | CCCEEEEECCEEEEEE | c1d-AT-4 | multi | 0.209 | 0.074 | 0.229 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.714 |
| 19 | fp_beta_D | P42224 | 331 | RPLVLKTGVQFTVKLRL | CCEEEEECCEEEEEEEE | c1c-4 | multi-selected | 0.181 | 0.063 | 0.194 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.75 |
| 20 | fp_beta_D | P42224 | 333 | LVLKTGVQFTVKLRLLV | EEEEECCEEEEEEEECC | c1c-AT-5 | multi | 0.155 | 0.051 | 0.15 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.75 |
| 21 | fp_beta_D | P42224 | 335 | LKTGVQFTVKLRLLVKL | EEECCEEEEEEEECCCC | c1c-4 | multi-selected | 0.139 | 0.046 | 0.116 | DISO | boundary|STAT_alpha; boundary|STAT_bind; | 0.875 |
| 22 | fp_beta_D | P42224 | 335 | LKTGVQFTVKLRLLV | EEECCEEEEEEEECC | c1b-AT-4 | multi | 0.141 | 0.047 | 0.125 | DISO | boundary|STAT_alpha; boundary|STAT_bind; | 0.857 |
| 23 | fp_beta_D | P42224 | 337 | TGVQFTVKLRLLVKLQE | ECCEEEEEEEECCCCCC | c1cR-4 | multi | 0.123 | 0.045 | 0.081 | DISO | boundary|STAT_alpha; boundary|STAT_bind; | 0.875 |
| 24 | fp_beta_D | P42224 | 337 | TGVQFTVKLRLLVKLQE | ECCEEEEEEEECCCCCC | c1c-4 | multi-selected | 0.123 | 0.045 | 0.081 | DISO | boundary|STAT_alpha; boundary|STAT_bind; | 0.875 |
| 25 | fp_beta_D | P42224 | 338 | GVQFTVKLRLLVKLQE | CCEEEEEEEECCCCCC | c1d-AT-5 | multi | 0.117 | 0.043 | 0.071 | DISO | boundary|STAT_alpha; MID|STAT_bind; | 0.857 |
| 26 | fp_beta_D | P42224 | 339 | VQFTVKLRLLVKLQELN | CEEEEEEEECCCCCCCC | c1cR-5 | multi | 0.122 | 0.045 | 0.061 | DISO | boundary|STAT_alpha; MID|STAT_bind; | 0.625 |
| 27 | fp_D | P42224 | 347 | LLVKLQELNYNLKVKV | ECCCCCCCCCCCCEEE | c1aR-4 | multi-selected | 0.174 | 0.052 | 0.077 | DISO | boundary|STAT_alpha; MID|STAT_bind; | 0.0 |
| 28 | fp_D | P42224 | 347 | LLVKLQELNYNLKVKV | ECCCCCCCCCCCCEEE | c1d-5 | multi | 0.174 | 0.052 | 0.077 | DISO | boundary|STAT_alpha; MID|STAT_bind; | 0.0 |
| 29 | fp_D | P42224 | 350 | KLQELNYNLKVKVLFDK | CCCCCCCCCCEEEEEEC | c1c-5 | multi-selected | 0.233 | 0.056 | 0.112 | DISO | boundary|STAT_alpha; MID|STAT_bind; | 0.25 |
| 30 | fp_D | P42224 | 369 | NERNTVKGFRKFNILG | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.688 | 0.214 | 0.246 | DISO | MID|STAT_bind; | 0.0 |
| 31 | fp_D | P42224 | 370 | ERNTVKGFRKFNILG | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.703 | 0.214 | 0.246 | DISO | MID|STAT_bind; | 0.0 |
| 32 | cand_D | P42224 | 379 | KFNILGTHTKVMNMEE +++ | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.49 | 0.204 | 0.337 | DISO | MID|STAT_bind; | 0.0 |
| 33 | cand_D | P42224 | 381 | NILGTHTKVMNMEE +++ | CCCCCCCCCCCCCC | c3-AT | multi | 0.459 | 0.208 | 0.345 | DISO | MID|STAT_bind; | 0.0 |
| 34 | cand_D | P42224 | 396 | TNGSLAAEFRHLQLKE ++++*+++*++*+*++ | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.191 | 0.287 | 0.365 | DISO | MID|STAT_bind; | 0.0 |
| 35 | cand_D | P42224 | 397 | NGSLAAEFRHLQLKE +++*+++*++*+*++ | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.192 | 0.287 | 0.36 | DISO | MID|STAT_bind; | 0.0 |
| 36 | cand_D | P42224 | 398 | GSLAAEFRHLQLKE ++*+++*++*+*++ | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.193 | 0.289 | 0.36 | DISO | MID|STAT_bind; | 0.0 |
| 37 | fp_D | P42224 | 422 | GPLIVTEELHSLSFET | CCCCCCCCEEEEEEEE | c1a-4 | multi-selected | 0.199 | 0.056 | 0.267 | boundary | MID|STAT_bind; | 0.429 |
| 38 | fp_beta_D | P42224 | 423 | PLIVTEELHSLSFET | CCCCCCCEEEEEEEE | c1b-AT-5 | multi | 0.168 | 0.041 | 0.25 | boundary | MID|STAT_bind; | 0.571 |
| 39 | fp_D | P42224 | 444 | LVIDLETTSLPVVV | CEEEECCCCCCEEE | c2-AT-5 | multi-selected | 0.051 | 0.019 | 0.136 | boundary | MID|STAT_bind; | 0.286 |
| 40 | fp_D | P42224 | 447 | DLETTSLPVVVISN | EECCCCCCEEEEEC | c3-AT | multi-selected | 0.076 | 0.017 | 0.134 | boundary | MID|STAT_bind; | 0.286 |
| 41 | fp_D | P42224 | 502 | LSWQFSSVTKRGLNVDQ | HHHHCCCCCCCCCCHHH | c1c-AT-4 | uniq | 0.032 | 0.051 | 0.109 | boundary | MID|STAT_bind; | 0.0 |
| 42 | fp_D | P42224 | 510 | TKRGLNVDQLNMLG | CCCCCCHHHHHHHH | c2-4 | uniq | 0.049 | 0.069 | 0.174 | boundary | MID|STAT_bind; | 0.0 |
| 43 | fp_O | P42224 | 550 | KNFPFWLWIESILELIK | CCCCHHHHHHHHHHHHH | c1c-4 | uniq | 0.012 | 0.016 | 0.019 | ORD | boundary|STAT_bind; boundary|SH2_STAT1; | 0.0 |
| 44 | fp_O | P42224 | 631 | VEPYTKKELSAVTFPD | CCCCCHHHHCCCCCHH | c1a-AT-4 | uniq | 0.022 | 0.048 | 0.267 | ORD | MID|SH2_STAT1; | 0.0 |
| 45 | fp_D | P42224 | 698 | GTGYIKTELISVSE | CCCCCCCCEEEECC | c2-AT-4 | uniq | 0.137 | 0.174 | 0.439 | DISO | boundary|SH2_STAT1; | 0.286 |
| 46 | fp_D | P42224 | 730 | EFDEVSRIVGSVEFDS | CCCCCCCCCCCCCCCC | c1a-5 | multi-selected | 0.953 | 0.359 | 0.366 | DISO | boundary|SH2_STAT1; | 0.0 |
| 47 | fp_D | P42224 | 730 | EFDEVSRIVGSVEFDS | CCCCCCCCCCCCCCCC | c1d-5 | multi | 0.953 | 0.359 | 0.366 | DISO | boundary|SH2_STAT1; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment