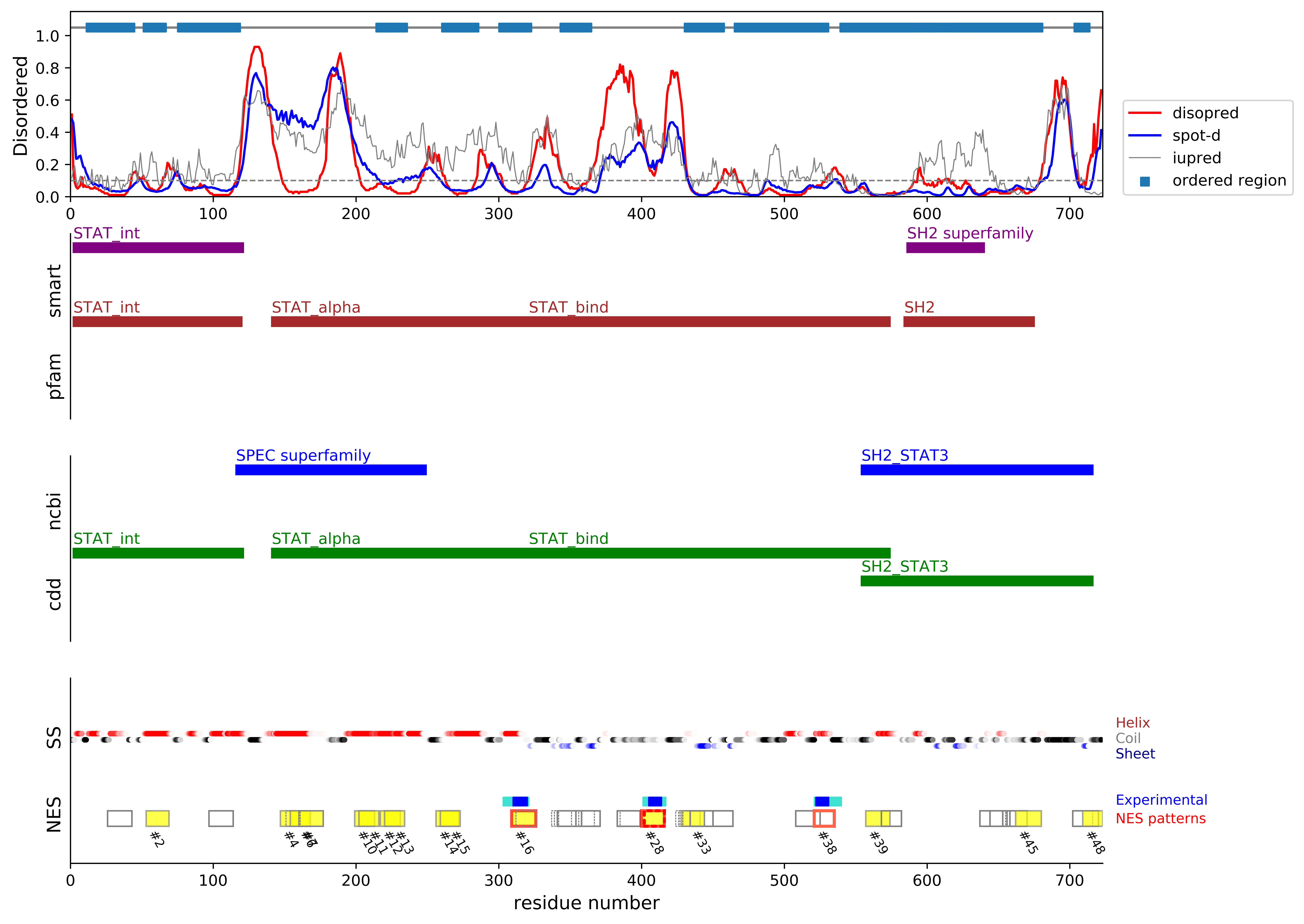

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P42227 | 26 | FPMELRQFLAPWIESQD | CCHHHHHHHHHHHHCCC | c1cR-4 | uniq | 0.018 | 0.038 | 0.083 | boundary | MID|STAT_int; | 0.0 |
| 2 | fp_D | P42227 | 53 | ATLVFHNLLGEIDQQY | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.079 | 0.039 | 0.156 | boundary | MID|STAT_int; | 0.0 |
| 3 | fp_D | P42227 | 97 | KPMEIARIVARCLWEES | CCHHHHHHHHHHHHHHH | c1cR-4 | uniq | 0.014 | 0.046 | 0.125 | boundary | boundary|STAT_int; | 0.0 |
| 4 | fp_D | P42227 | 147 | HLQDVRKRVQDLEQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.051 | 0.493 | 0.41 | DISO | boundary|STAT_int; boundary|STAT_alpha; | 0.0 |
| 5 | fp_D | P42227 | 151 | VRKRVQDLEQKMKVVE | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.031 | 0.477 | 0.373 | DISO | boundary|STAT_int; boundary|STAT_alpha; | 0.0 |
| 6 | fp_D | P42227 | 154 | RVQDLEQKMKVVEN | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.029 | 0.474 | 0.356 | DISO | boundary|STAT_int; boundary|STAT_alpha; | 0.0 |
| 7 | fp_D | P42227 | 160 | QKMKVVENLQDDFDFNY | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.052 | 0.467 | 0.324 | DISO | boundary|STAT_alpha; | 0.0 |
| 8 | fp_D | P42227 | 160 | QKMKVVENLQDDFDFNY | HHHHHHHHHHHHHHHHH | c1cR-4 | multi | 0.052 | 0.467 | 0.324 | DISO | boundary|STAT_alpha; | 0.0 |
| 9 | fp_D | P42227 | 161 | KMKVVENLQDDFDFNY | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.054 | 0.467 | 0.319 | DISO | boundary|STAT_alpha; | 0.0 |
| 10 | fp_D | P42227 | 199 | KMQQLEQMLTALDQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.094 | 0.216 | 0.43 | boundary | MID|STAT_alpha; | 0.0 |
| 11 | fp_D | P42227 | 202 | QLEQMLTALDQMRR | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.064 | 0.176 | 0.381 | boundary | MID|STAT_alpha; | 0.0 |
| 12 | fp_D | P42227 | 217 | IVSELAGLLSAMEY | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.05 | 0.089 | 0.244 | boundary | MID|STAT_alpha; | 0.0 |
| 13 | fp_D | P42227 | 220 | ELAGLLSAMEYVQK | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.037 | 0.092 | 0.245 | boundary | MID|STAT_alpha; | 0.0 |
| 14 | fp_D | P42227 | 256 | PNICLDRLENWITSLA | CCCCCCCHHHHHHHHH | c1aR-5 | multi-selected | 0.109 | 0.071 | 0.218 | boundary | MID|STAT_alpha; | 0.0 |
| 15 | fp_D | P42227 | 259 | CLDRLENWITSLAE | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.073 | 0.057 | 0.23 | boundary | MID|STAT_alpha; | 0.0 |
| 16 | cand_D | P42227 | 309 | IVELFRNLMKSAFVVER ++++*+++*+ | HHHHHHHHHCCCEEEEE | c1c-5 | multi-selected | 0.081 | 0.043 | 0.188 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.0 |
| 17 | fp_D | P42227 | 312 | LFRNLMKSAFVVER +*+++*+ | HHHHHHCCCEEEEE | c3-AT | multi | 0.094 | 0.045 | 0.178 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.143 |
| 18 | fp_beta_D | P42227 | 337 | LVIKTGVQFTTKVRLLV | CEEEECCEEEEEEEEEC | c1c-AT-5 | multi | 0.145 | 0.038 | 0.206 | __ | boundary|STAT_alpha; boundary|STAT_bind; | 0.75 |
| 19 | fp_beta_D | P42227 | 339 | IKTGVQFTTKVRLLVKF | EEECCEEEEEEEEECCC | c1c-AT-4 | multi | 0.117 | 0.033 | 0.17 | __ | boundary|STAT_alpha; boundary|STAT_bind; | 0.875 |
| 20 | fp_beta_D | P42227 | 339 | IKTGVQFTTKVRLLV | EEECCEEEEEEEEEC | c1b-AT-4 | multi | 0.126 | 0.033 | 0.183 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 0.857 |
| 21 | fp_beta_D | P42227 | 341 | TGVQFTTKVRLLVKFPE | ECCEEEEEEEEECCCCC | c1cR-4 | multi | 0.094 | 0.031 | 0.131 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 1.0 |
| 22 | fp_beta_D | P42227 | 341 | TGVQFTTKVRLLVKFPE | ECCEEEEEEEEECCCCC | c1c-4 | multi-selected | 0.094 | 0.031 | 0.131 | boundary | boundary|STAT_alpha; boundary|STAT_bind; | 1.0 |
| 23 | fp_beta_D | P42227 | 342 | GVQFTTKVRLLVKFPE | CCEEEEEEEEECCCCC | c1d-AT-5 | multi | 0.083 | 0.029 | 0.12 | boundary | boundary|STAT_alpha; MID|STAT_bind; | 1.0 |
| 24 | fp_D | P42227 | 351 | LLVKFPELNYQLKIKV | EECCCCCCCCEEEEEE | c1d-5 | multi | 0.083 | 0.036 | 0.093 | boundary | boundary|STAT_alpha; MID|STAT_bind; | 0.143 |
| 25 | fp_beta_D | P42227 | 354 | KFPELNYQLKIKVCIDK | CCCCCCCEEEEEEEEEC | c1c-5 | multi-selected | 0.13 | 0.04 | 0.098 | boundary | boundary|STAT_alpha; MID|STAT_bind; | 0.625 |
| 26 | fp_D | P42227 | 383 | KFNILGTNTKVMNMEE | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.685 | 0.256 | 0.371 | DISO | MID|STAT_bind; | 0.0 |
| 27 | fp_D | P42227 | 385 | NILGTNTKVMNMEE | CCCCCCCCCCCCCC | c3-AT | multi | 0.673 | 0.262 | 0.39 | DISO | MID|STAT_bind; | 0.0 |
| 28 | cand_D | P42227 | 400 | NNGSLSAEFKHLTLRE ++++*++*+++ | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.232 | 0.23 | 0.366 | DISO | MID|STAT_bind; | 0.0 |
| 29 | cand_D | P42227 | 402 | GSLSAEFKHLTLRE ++++*++*+++ | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.207 | 0.219 | 0.357 | DISO | MID|STAT_bind; | 0.0 |
| 30 | fp_D | P42227 | 424 | ANCDASLIVTEELHLIT | CCCCCCCHHHHHHHEEE | c1c-AT-4 | multi | 0.289 | 0.175 | 0.17 | boundary | MID|STAT_bind; | 0.0 |
| 31 | fp_D | P42227 | 426 | CDASLIVTEELHLIT | CCCCCHHHHHHHEEE | c1b-AT-4 | multi | 0.225 | 0.14 | 0.143 | boundary | MID|STAT_bind; | 0.0 |
| 32 | fp_D | P42227 | 427 | DASLIVTEELHLIT | CCCCHHHHHHHEEE | c2-AT-4 | multi | 0.19 | 0.125 | 0.133 | boundary | MID|STAT_bind; | 0.0 |
| 33 | fp_D | P42227 | 428 | ASLIVTEELHLITFET | CCCHHHHHHHEEEEEE | c1a-4 | multi-selected | 0.128 | 0.088 | 0.122 | boundary | MID|STAT_bind; | 0.143 |
| 34 | fp_D | P42227 | 429 | SLIVTEELHLITFET | CCHHHHHHHEEEEEE | c1b-AT-5 | multi | 0.098 | 0.072 | 0.117 | boundary | MID|STAT_bind; | 0.286 |
| 35 | fp_beta_D | P42227 | 434 | EELHLITFETEVYHQG | HHHHEEEEEEEEEECC | c1aR-4 | multi-selected | 0.021 | 0.021 | 0.108 | __ | MID|STAT_bind; | 1.0 |
| 36 | fp_D | P42227 | 450 | LKIDLETHSLPVVV | CEEEECCCCCCCEE | c2-AT-4 | uniq | 0.109 | 0.03 | 0.138 | boundary | MID|STAT_bind; | 0.286 |
| 37 | fp_D | P42227 | 508 | LSWQFSSTTKRGLSIEQ + | HHHHHHHHCCCCCCHHH | c1c-AT-4 | uniq | 0.039 | 0.048 | 0.14 | boundary | MID|STAT_bind; | 0.0 |
| 38 | cand_D | P42227 | 521 | SIEQLTTLAEKLLG +*++*++++++ | CHHHHHHHHHHHCC | c3-AT | uniq | 0.101 | 0.064 | 0.155 | boundary | MID|STAT_bind; boundary|SH2_STAT3; | 0.0 |
| 39 | fp_D | P42227 | 557 | KGFSFWVWLDNIIDLVK | CCCCCHHHHHHHHHHHH | c1c-4 | uniq | 0.015 | 0.02 | 0.016 | boundary | boundary|STAT_bind; boundary|SH2_STAT3; | 0.0 |
| 40 | fp_O | P42227 | 568 | IIDLVKKYILALWN | HHHHHHHHHHHHHC | c3-4 | uniq | 0.018 | 0.01 | 0.009 | ORD | boundary|STAT_bind; boundary|SH2_STAT3; | 0.0 |
| 41 | fp_O | P42227 | 637 | VEPYTKQQLNNMSFAE | ECCCCHHHCCCCCHHH | c1a-AT-4 | uniq | 0.021 | 0.036 | 0.2 | ORD | MID|SH2_STAT3; | 0.0 |
| 42 | fp_O | P42227 | 644 | QLNNMSFAEIIMGY | HCCCCCHHHHHCCC | c2-5 | uniq | 0.025 | 0.039 | 0.089 | ORD | MID|SH2_STAT3; | 0.0 |
| 43 | fp_D | P42227 | 655 | MGYKIMDATNILVSP | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.033 | 0.051 | 0.031 | boundary | MID|SH2_STAT3; | 0.0 |
| 44 | fp_D | P42227 | 656 | GYKIMDATNILVSP | CCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.034 | 0.052 | 0.03 | boundary | MID|SH2_STAT3; | 0.0 |
| 45 | fp_D | P42227 | 662 | ATNILVSPLVYLYPDIPK | CCCCCCCCCCCCCCCCCH | c4-4 | uniq | 0.044 | 0.057 | 0.055 | boundary | MID|SH2_STAT3; | 0.0 |
| 46 | fp_beta_D | P42227 | 702 | AAPYLKTKFICVTPFIDA | CCCCCCCEEEEEECCCCC | c4-4 | multi-selected | 0.188 | 0.118 | 0.103 | __ | boundary|SH2_STAT3; | 0.75 |
| 47 | fp_D | P42227 | 702 | AAPYLKTKFICVTP | CCCCCCCEEEEEEC | c2-AT-4 | multi | 0.148 | 0.097 | 0.126 | __ | boundary|SH2_STAT3; | 0.429 |
| 48 | fp_D | P42227 | 709 | KFICVTPFIDAVWK | EEEEEECCCCCCCC | c3-4 | uniq | 0.291 | 0.154 | 0.029 | boundary | boundary|SH2_STAT3; | 0.429 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment