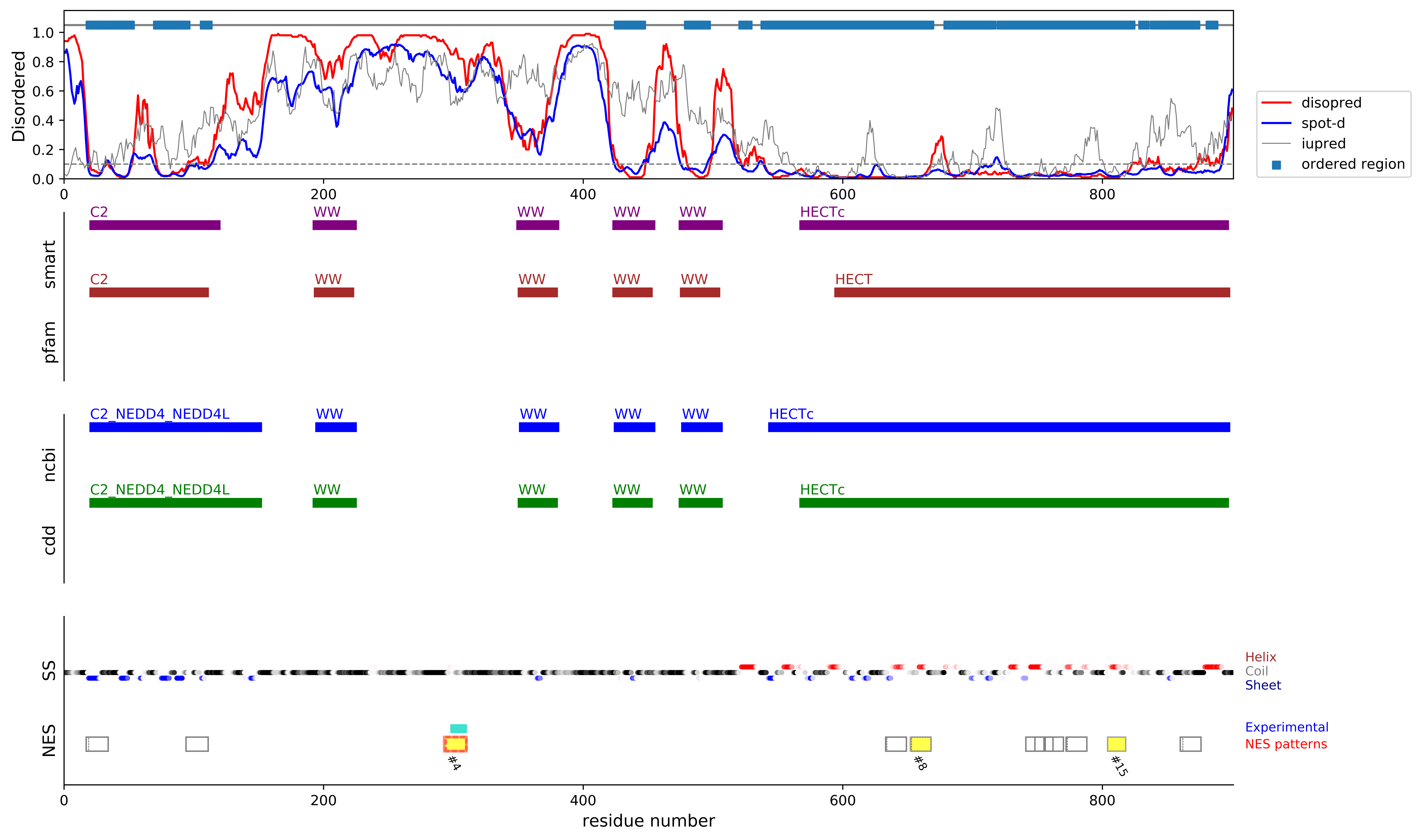

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | P46934-4 | 17 | NSRIVRVRVIAGIGLAK | CCEEEEEEEEEECCCCC | c1c-4 | multi-selected | 0.101 | 0.065 | 0.109 | boundary | boundary|C2_NEDD4_NEDD4L; | 1.0 |
| 2 | fp_beta_D | P46934-4 | 19 | RIVRVRVIAGIGLAK | EEEEEEEEEECCCCC | c1b-5 | multi | 0.069 | 0.048 | 0.112 | boundary | boundary|C2_NEDD4_NEDD4L; | 0.857 |
| 3 | fp_D | P46934-4 | 94 | ENRLTRDDFLGQVDVPL | CCCCCCCCCCCEEEECC | c1c-AT-4 | uniq | 0.115 | 0.079 | 0.292 | boundary | MID|C2_NEDD4_NEDD4L; | 0.125 |
| 4 | cand_D | P46934-4 | 293 | VPTHLAEELNARLTIFG +++++++ | CCCCHHHHCCCCCCCCC | c1c-4 | multi-selected | 0.854 | 0.649 | 0.609 | DISO | 0.0 | |
| 5 | cand_D | P46934-4 | 294 | PTHLAEELNARLTIFG +++++++ | CCCHHHHCCCCCCCCC | c1d-AT-4 | multi | 0.849 | 0.646 | 0.594 | DISO | 0.0 | |
| 6 | fp_O | P46934-4 | 633 | SYFKFIGRVAGMAVYH | CCEEEHHHHHHHHHHH | c1a-4 | multi-selected | 0.01 | 0.01 | 0.022 | ORD | MID|HECTc; | 0.143 |
| 7 | fp_O | P46934-4 | 634 | YFKFIGRVAGMAVYH | CEEEHHHHHHHHHHH | c1b-5 | multi | 0.01 | 0.01 | 0.02 | ORD | MID|HECTc; | 0.0 |
| 8 | fp_D | P46934-4 | 652 | LDGFFIRPFYKMMLHK | CCCCCCHHHHHHHHCC | c1a-4 | multi-selected | 0.025 | 0.02 | 0.016 | boundary | MID|HECTc; | 0.0 |
| 9 | fp_D | P46934-4 | 653 | DGFFIRPFYKMMLHK | CCCCCHHHHHHHHCC | c1b-4 | multi | 0.026 | 0.021 | 0.016 | boundary | MID|HECTc; | 0.0 |
| 10 | fp_O | P46934-4 | 741 | FVNRIQKQMAAFKE | ECHHHHHHHHHHHH | c3-4 | uniq | 0.026 | 0.026 | 0.042 | ORD | MID|HECTc; | 0.0 |
| 11 | fp_O | P46934-4 | 748 | QMAAFKEGFFELIP | HHHHHHHHCCCCCC | c3-4 | uniq | 0.026 | 0.022 | 0.053 | ORD | MID|HECTc; | 0.0 |
| 12 | fp_O | P46934-4 | 756 | FFELIPQDLIKIFD | CCCCCCCHHHCCCC | c3-4 | uniq | 0.03 | 0.022 | 0.049 | ORD | MID|HECTc; | 0.0 |
| 13 | fp_O | P46934-4 | 772 | ELELLMCGLGDVDVND | HHHHHHHCCCCCCHHH | c1a-5 | multi-selected | 0.021 | 0.026 | 0.124 | ORD | MID|HECTc; | 0.0 |
| 14 | fp_O | P46934-4 | 773 | LELLMCGLGDVDVND | HHHHHHCCCCCCHHH | c1b-4 | multi | 0.02 | 0.027 | 0.127 | ORD | MID|HECTc; | 0.0 |
| 15 | fp_D | P46934-4 | 804 | VIQWFWKAVLMMDS | HHHHHHHHHHCCCH | c3-4 | uniq | 0.017 | 0.024 | 0.043 | boundary | MID|HECTc; | 0.0 |
| 16 | fp_D | P46934-4 | 860 | KLPRAHTCFNRLDLPP | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.08 | 0.049 | 0.273 | boundary | boundary|HECTc; | 0.0 |
| 17 | fp_D | P46934-4 | 862 | PRAHTCFNRLDLPP | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.082 | 0.046 | 0.268 | boundary | boundary|HECTc; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment