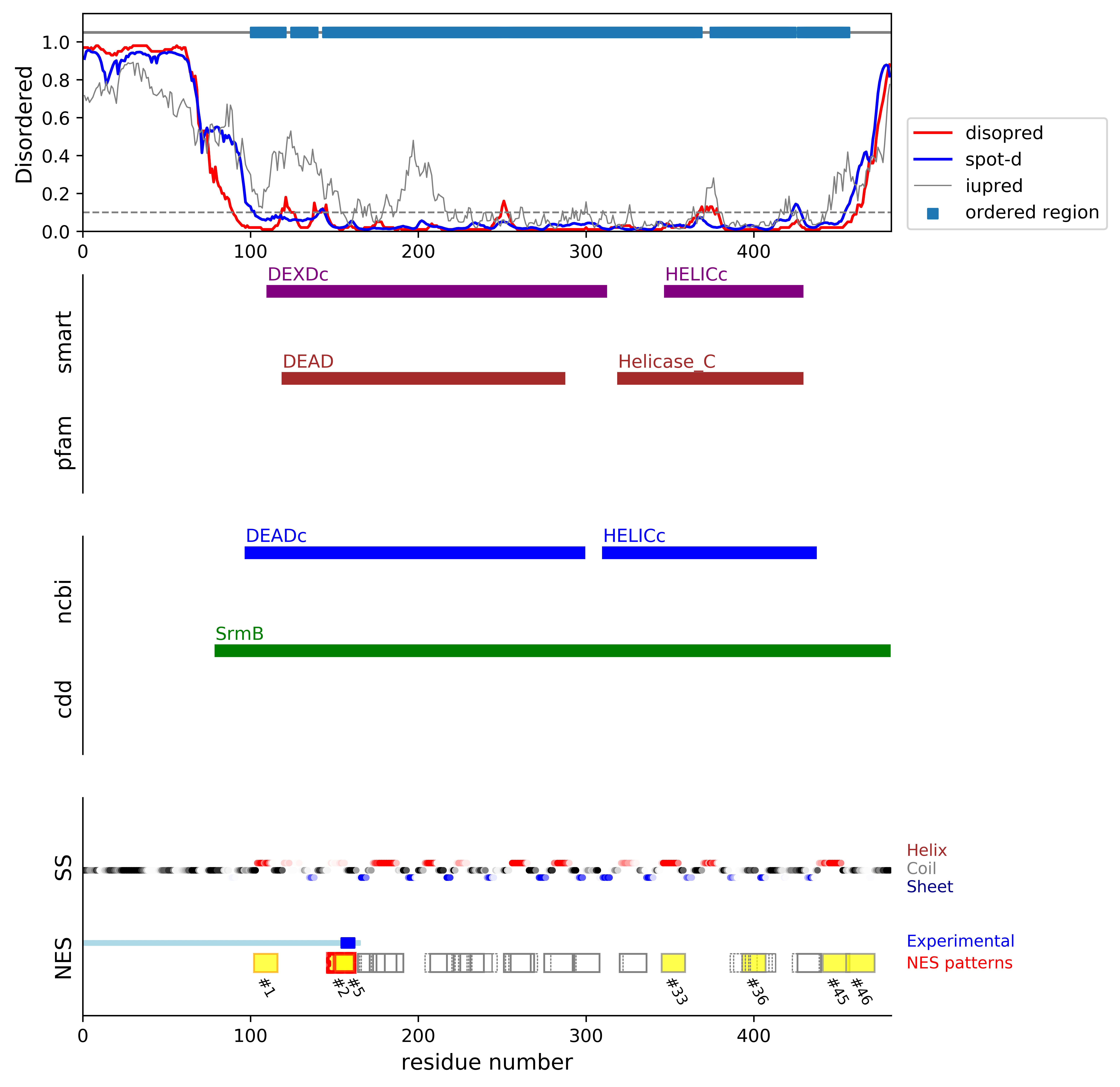

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P54824 | 102 | LKRELLMGIFEMGW .............. | CCHHHHHHHHHHCC | c2-4 | uniq | 0.016 | 0.072 | 0.247 | boundary | MID|SrmB; | 0.0 |
| 2 | cand_D | P54824 | 146 | SGAYLIPLLERLDLKK ...........*.*.. | CHHHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.028 | 0.032 | 0.142 | boundary | MID|SrmB; | 0.0 |
| 3 | cand_D | P54824 | 146 | SGAYLIPLLERLDLKK ...........*.*.. | CHHHHHHHHHHCCCCC | c1d-4 | multi | 0.028 | 0.032 | 0.142 | boundary | MID|SrmB; | 0.0 |
| 4 | cand_D | P54824 | 147 | GAYLIPLLERLDLKK ..........*.*.. | HHHHHHHHHHCCCCC | c1b-4 | multi | 0.025 | 0.03 | 0.135 | boundary | MID|SrmB; | 0.0 |
| 5 | cand_D | P54824 | 150 | LIPLLERLDLKK .......*.*.. | HHHHHHHCCCCC | c2-rev | multi-selected | 0.02 | 0.03 | 0.109 | boundary | MID|SrmB; | 0.0 |
| 6 | fp_O | P54824 | 164 | IQAMVIVPTRELALQV . | EEEEEECCCHHHHHHH | c1a-AT-4 | multi-selected | 0.027 | 0.014 | 0.099 | ORD | MID|SrmB; | 0.286 |
| 7 | fp_O | P54824 | 165 | QAMVIVPTRELALQV | EEEEECCCHHHHHHH | c1b-AT-4 | multi | 0.028 | 0.014 | 0.094 | ORD | MID|SrmB; | 0.143 |
| 8 | fp_O | P54824 | 166 | AMVIVPTRELALQV | EEEECCCHHHHHHH | c2-AT-5 | multi | 0.029 | 0.014 | 0.089 | ORD | MID|SrmB; | 0.143 |
| 9 | fp_O | P54824 | 171 | PTRELALQVSQICIQV | CCHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.029 | 0.015 | 0.103 | ORD | MID|SrmB; | 0.0 |
| 10 | fp_O | P54824 | 172 | TRELALQVSQICIQV | CHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.029 | 0.015 | 0.1 | ORD | MID|SrmB; | 0.0 |
| 11 | fp_O | P54824 | 173 | RELALQVSQICIQV | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.03 | 0.015 | 0.101 | ORD | MID|SrmB; | 0.0 |
| 12 | fp_O | P54824 | 175 | LALQVSQICIQVSKHM | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.026 | 0.016 | 0.123 | ORD | MID|SrmB; | 0.0 |
| 13 | fp_O | P54824 | 204 | LRDDIMRLDDTVHVVI | HHHHHHHHCCCCEEEE | c1d-4 | multi | 0.019 | 0.023 | 0.201 | ORD | MID|SrmB; | 0.0 |
| 14 | fp_O | P54824 | 207 | DIMRLDDTVHVVIA | HHHHHCCCCEEEEC | c3-4 | multi-selected | 0.019 | 0.019 | 0.163 | ORD | MID|SrmB; | 0.143 |
| 15 | fp_O | P54824 | 207 | DIMRLDDTVHVVIAT | HHHHHCCCCEEEECC | c1b-AT-5 | multi | 0.019 | 0.019 | 0.157 | ORD | MID|SrmB; | 0.286 |
| 16 | fp_O | P54824 | 207 | DIMRLDDTVHVVIATPG | HHHHHCCCCEEEECCCH | c1cR-4 | multi | 0.018 | 0.018 | 0.149 | ORD | MID|SrmB; | 0.375 |
| 17 | fp_O | P54824 | 217 | VVIATPGRILDLIK | EEECCCHHHHHHHC | c3-AT | uniq | 0.01 | 0.014 | 0.073 | ORD | MID|SrmB; | 0.0 |
| 18 | fp_O | P54824 | 225 | ILDLIKKGVAKVDH | HHHHHCCCCCCCCC | c3-4 | multi-selected | 0.01 | 0.029 | 0.059 | ORD | MID|SrmB; | 0.0 |
| 19 | fp_O | P54824 | 229 | IKKGVAKVDHIQMIV | HCCCCCCCCCCCEEE | c1b-4 | multi | 0.01 | 0.03 | 0.064 | ORD | MID|SrmB; | 0.0 |
| 20 | fp_O | P54824 | 230 | KKGVAKVDHIQMIV | CCCCCCCCCCCEEE | c2-AT-4 | multi | 0.01 | 0.031 | 0.064 | ORD | MID|SrmB; | 0.0 |
| 21 | fp_O | P54824 | 232 | GVAKVDHIQMIVLDE | CCCCCCCCCEEEEEC | c1b-5 | multi | 0.017 | 0.029 | 0.057 | ORD | MID|SrmB; | 0.286 |
| 22 | fp_O | P54824 | 251 | LSQDFMQIMEDIIMTL | CCCCHHHHHHHHHHHC | c1a-4 | multi-selected | 0.044 | 0.026 | 0.064 | ORD | MID|SrmB; | 0.0 |
| 23 | fp_O | P54824 | 251 | LSQDFMQIMEDIIMTL | CCCCHHHHHHHHHHHC | c1d-4 | multi | 0.044 | 0.026 | 0.064 | ORD | MID|SrmB; | 0.0 |
| 24 | fp_O | P54824 | 252 | SQDFMQIMEDIIMTL | CCCHHHHHHHHHHHC | c1b-4 | multi | 0.036 | 0.024 | 0.064 | ORD | MID|SrmB; | 0.0 |
| 25 | fp_O | P54824 | 254 | DFMQIMEDIIMTLPKNR | CHHHHHHHHHHHCCCCC | c1cR-4 | multi | 0.021 | 0.02 | 0.063 | ORD | MID|SrmB; | 0.0 |
| 26 | fp_O | P54824 | 255 | FMQIMEDIIMTLPK | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.018 | 0.019 | 0.066 | ORD | MID|SrmB; | 0.0 |
| 27 | fp_O | P54824 | 275 | YSATFPLSVQKFMTLHL | EECCCCHHHHHHHHHHC | c1c-4 | multi-selected | 0.01 | 0.031 | 0.052 | ORD | MID|SrmB; | 0.0 |
| 28 | fp_O | P54824 | 279 | FPLSVQKFMTLHLQK | CCHHHHHHHHHHCCC | c1b-4 | multi | 0.01 | 0.035 | 0.056 | ORD | MID|SrmB; | 0.0 |
| 29 | fp_O | P54824 | 293 | KPYEINLMEELTLKG | CCEEEECCCCCCCCC | c1b-4 | uniq | 0.011 | 0.039 | 0.075 | ORD | MID|SrmB; | 0.286 |
| 30 | fp_O | P54824 | 320 | KVHCLNTLFSRLQINQ | HHHHHHHHHHHCCCCC | c1a-5 | multi-selected | 0.02 | 0.013 | 0.042 | ORD | MID|SrmB; | 0.0 |
| 31 | fp_O | P54824 | 320 | KVHCLNTLFSRLQINQ | HHHHHHHHHHHCCCCC | c1d-5 | multi | 0.02 | 0.013 | 0.042 | ORD | MID|SrmB; | 0.0 |
| 32 | fp_O | P54824 | 322 | HCLNTLFSRLQINQ | HHHHHHHHHCCCCC | c2-AT-4 | multi | 0.019 | 0.013 | 0.044 | ORD | MID|SrmB; | 0.0 |
| 33 | fp_D | P54824 | 345 | RVELLAKKISQLGY | HHHHHHHHHHHCCC | c3-4 | uniq | 0.025 | 0.032 | 0.028 | boundary | MID|SrmB; | 0.0 |
| 34 | fp_D | P54824 | 386 | NLVCTDLFTRGIDIQA | EEEEECCCCCCCCCCC | c1d-AT-5 | multi | 0.01 | 0.021 | 0.034 | boundary | MID|SrmB; | 0.143 |
| 35 | fp_D | P54824 | 388 | VCTDLFTRGIDIQA | EEECCCCCCCCCCC | c2-AT-4 | multi | 0.01 | 0.022 | 0.033 | boundary | MID|SrmB; | 0.0 |
| 36 | fp_D | P54824 | 393 | FTRGIDIQAVNVVI | CCCCCCCCCEEEEE | c2-4 | multi-selected | 0.009 | 0.023 | 0.033 | boundary | MID|SrmB; | 0.143 |
| 37 | fp_D | P54824 | 393 | FTRGIDIQAVNVVINF | CCCCCCCCCEEEEEEC | c1a-AT-4 | multi | 0.009 | 0.022 | 0.033 | boundary | MID|SrmB; | 0.286 |
| 38 | fp_beta_D | P54824 | 395 | RGIDIQAVNVVINFDF | CCCCCCCEEEEEECCC | c1d-4 | multi | 0.009 | 0.021 | 0.034 | boundary | MID|SrmB; | 0.571 |
| 39 | fp_D | P54824 | 395 | RGIDIQAVNVVINF | CCCCCCCEEEEEEC | c2-AT-4 | multi | 0.009 | 0.022 | 0.033 | boundary | MID|SrmB; | 0.429 |
| 40 | fp_beta_D | P54824 | 397 | IDIQAVNVVINFDFPK | CCCCCEEEEEECCCCC | c1a-AT-4 | multi | 0.009 | 0.021 | 0.034 | boundary | MID|SrmB; | 0.857 |
| 41 | fp_beta_D | P54824 | 397 | IDIQAVNVVINFDFPK | CCCCCEEEEEECCCCC | c1d-AT-4 | multi | 0.009 | 0.021 | 0.034 | boundary | MID|SrmB; | 0.857 |
| 42 | fp_beta_D | P54824 | 398 | DIQAVNVVINFDFPK | CCCCEEEEEECCCCC | c1b-5 | multi | 0.009 | 0.02 | 0.035 | boundary | MID|SrmB; | 0.857 |
| 43 | fp_D | P54824 | 423 | RSGRFGHLGLAINLIT | CCCCCCCCCEEEEECC | c1d-4 | multi | 0.024 | 0.067 | 0.062 | boundary | MID|SrmB; | 0.286 |
| 44 | fp_beta_D | P54824 | 426 | RFGHLGLAINLITY | CCCCCCEEEEECCH | c3-4 | multi-selected | 0.019 | 0.052 | 0.054 | boundary | MID|SrmB; | 0.571 |
| 45 | fp_D | P54824 | 441 | DRFNLKSIEEQLGTEI | HHHHHHHHHHHHCCCC | c1aR-4 | uniq | 0.024 | 0.058 | 0.22 | boundary | MID|SrmB; | 0.0 |
| 46 | fp_D | P54824 | 455 | EIKPIPSSIDKNLYVAE | CCCCCCCCCCCCCCEEC | c1c-5 | uniq | 0.171 | 0.3 | 0.294 | boundary | boundary|SrmB; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment