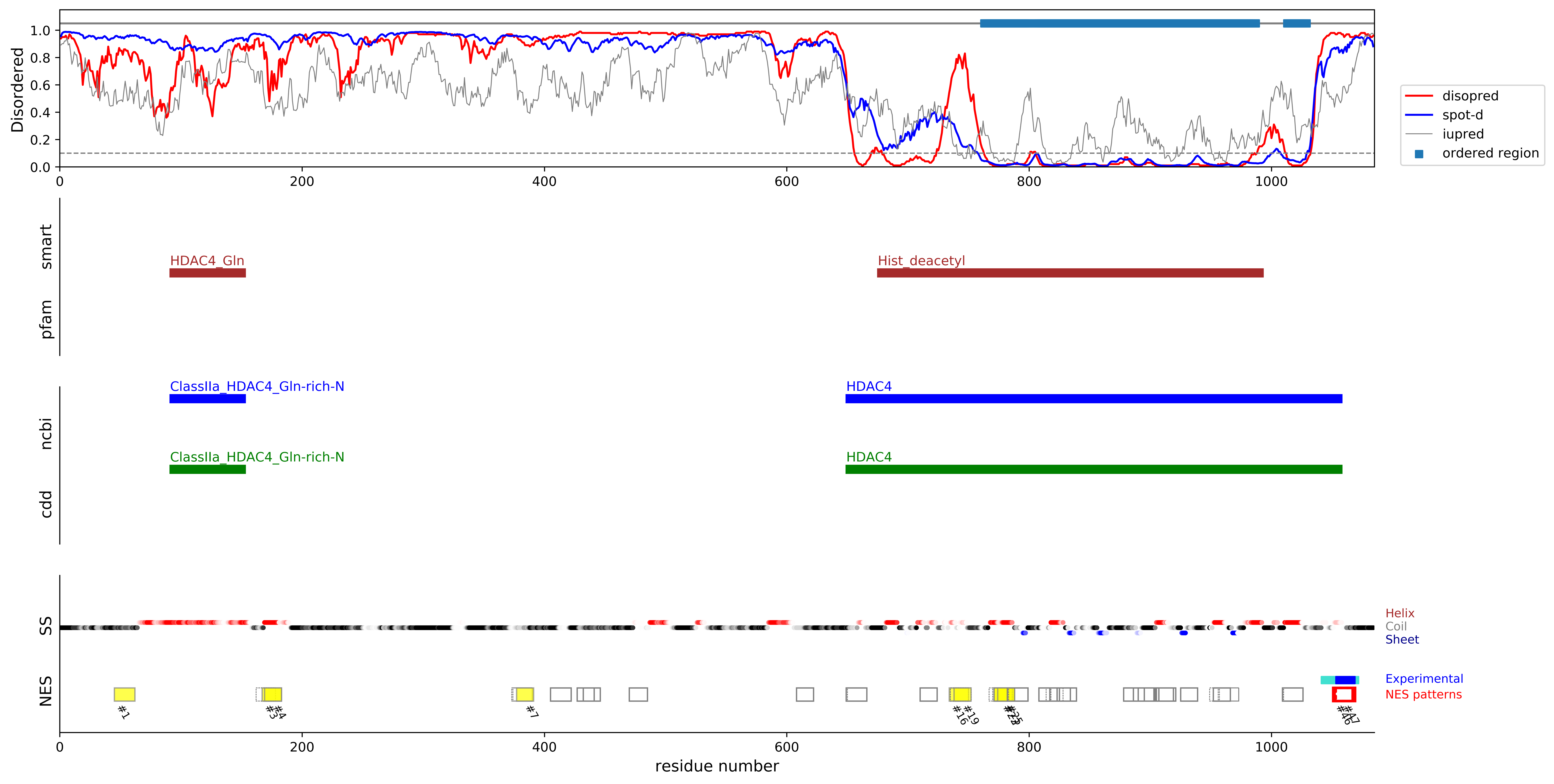

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P56524 | 45 | SAVPMDLRLDHQFSLPV | CCCCCCCCCCCCCCCCC | c1c-4 | uniq | 0.824 | 0.953 | 0.521 | DISO | 0.0 | |
| 2 | fp_D | P56524 | 162 | KESAVASTEVKMKLQE | CCCCCCCHHHHHHHHH | c1d-AT-4 | multi | 0.776 | 0.957 | 0.531 | DISO | boundary|ClassIIa_HDAC4_Gln-rich-N; | 0.0 |
| 3 | fp_D | P56524 | 167 | ASTEVKMKLQEFVLNK | CCHHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.702 | 0.93 | 0.468 | DISO | boundary|ClassIIa_HDAC4_Gln-rich-N; | 0.0 |
| 4 | fp_D | P56524 | 169 | TEVKMKLQEFVLNK | HHHHHHHHHHHHHC | c2-4 | multi-selected | 0.674 | 0.925 | 0.445 | DISO | boundary|ClassIIa_HDAC4_Gln-rich-N; | 0.0 |
| 5 | fp_D | P56524 | 373 | AERLTLPALQQRLSLFP | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi | 0.922 | 0.954 | 0.549 | DISO | 0.0 | |
| 6 | fp_D | P56524 | 374 | ERLTLPALQQRLSLFP | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.919 | 0.954 | 0.534 | DISO | 0.0 | |
| 7 | fp_D | P56524 | 377 | TLPALQQRLSLFPG | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.909 | 0.951 | 0.488 | DISO | 0.0 | |
| 8 | fp_D | P56524 | 405 | DGGAAHSPLLQHMVLLE | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.955 | 0.899 | 0.593 | DISO | 0.0 | |
| 9 | fp_D | P56524 | 427 | APLVTGLGALPLHA | CCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.981 | 0.904 | 0.503 | DISO | 0.0 | |
| 10 | fp_D | P56524 | 432 | GLGALPLHAQSLVG | CCCCCCCCCCCCCC | c3-AT | multi-selected | 0.98 | 0.88 | 0.495 | DISO | 0.0 | |
| 11 | fp_D | P56524 | 470 | LPQNAQALQHLVIQQ | CCCCHHHHHCHHCCC | c1b-AT-4 | uniq | 0.981 | 0.897 | 0.637 | DISO | 0.0 | |
| 12 | fp_D | P56524 | 608 | YQASMEAAGIPVSF | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.925 | 0.897 | 0.57 | DISO | 0.0 | |
| 13 | fp_D | P56524 | 649 | KPRFTTGLVYDTLMLKH | CCCCCCCCCCHHHHHHC | c1c-AT-4 | multi-selected | 0.174 | 0.457 | 0.395 | DISO | boundary|HDAC4; | 0.0 |
| 14 | fp_D | P56524 | 650 | PRFTTGLVYDTLMLKH | CCCCCCCCCHHHHHHC | c1d-AT-4 | multi | 0.141 | 0.447 | 0.388 | DISO | boundary|HDAC4; | 0.0 |
| 15 | fp_D | P56524 | 710 | ELQTVHSEAHTLLY | HHHHCCCHHHHHHH | c3-AT | uniq | 0.05 | 0.356 | 0.402 | DISO | MID|HDAC4; | 0.0 |
| 16 | fp_D | P56524 | 734 | DSKKLLGSLASVFVRL | HHHHHHCCCHHHHHHC | c1a-4 | multi-selected | 0.725 | 0.231 | 0.147 | boundary | MID|HDAC4; | 0.0 |
| 17 | fp_D | P56524 | 735 | SKKLLGSLASVFVRL | HHHHHCCCHHHHHHC | c1b-4 | multi | 0.737 | 0.224 | 0.13 | boundary | MID|HDAC4; | 0.0 |
| 18 | fp_D | P56524 | 735 | SKKLLGSLASVFVRLPC | HHHHHCCCHHHHHHCCC | c1c-AT-4 | multi | 0.716 | 0.216 | 0.13 | boundary | MID|HDAC4; | 0.0 |
| 19 | fp_D | P56524 | 738 | LLGSLASVFVRLPC | HHCCCHHHHHHCCC | c3-4 | multi-selected | 0.732 | 0.193 | 0.104 | boundary | MID|HDAC4; | 0.0 |
| 20 | fp_D | P56524 | 767 | SAGAARLAVGCVVELVF | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.014 | 0.017 | 0.073 | boundary | MID|HDAC4; | 0.0 |
| 21 | fp_D | P56524 | 770 | AARLAVGCVVELVFKV | HHHHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.012 | 0.014 | 0.056 | boundary | MID|HDAC4; | 0.0 |
| 22 | fp_D | P56524 | 771 | ARLAVGCVVELVFKVA | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.013 | 0.014 | 0.052 | __ | MID|HDAC4; | 0.0 |
| 23 | fp_D | P56524 | 771 | ARLAVGCVVELVFKVAT | HHHHHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.014 | 0.015 | 0.051 | __ | MID|HDAC4; | 0.0 |
| 24 | fp_D | P56524 | 771 | ARLAVGCVVELVFKV | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.013 | 0.014 | 0.053 | __ | MID|HDAC4; | 0.0 |
| 25 | fp_D | P56524 | 774 | AVGCVVELVFKVAT | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.014 | 0.014 | 0.048 | boundary | MID|HDAC4; | 0.0 |
| 26 | fp_O | P56524 | 782 | VFKVATGELKNGFAVVR | HHHHHCCCCCCCEEEEC | c1c-AT-5 | multi-selected | 0.022 | 0.018 | 0.211 | ORD | MID|HDAC4; | 0.0 |
| 27 | fp_O | P56524 | 783 | FKVATGELKNGFAVVR | HHHHCCCCCCCEEEEC | c1d-AT-4 | multi | 0.022 | 0.018 | 0.22 | ORD | MID|HDAC4; | 0.0 |
| 28 | fp_O | P56524 | 808 | TPMGFCYFNSVAVAA | CCCCCCCCCCHHHHH | c1b-4 | uniq | 0.015 | 0.024 | 0.117 | ORD | MID|HDAC4; | 0.0 |
| 29 | fp_O | P56524 | 814 | YFNSVAVAAKLLQQ | CCCCHHHHHHHHHH | c3-AT | multi | 0.015 | 0.013 | 0.045 | ORD | MID|HDAC4; | 0.0 |
| 30 | fp_O | P56524 | 817 | SVAVAAKLLQQRLSVSK | CHHHHHHHHHHHCCCCE | c1c-AT-5 | multi-selected | 0.016 | 0.013 | 0.039 | ORD | MID|HDAC4; | 0.0 |
| 31 | fp_O | P56524 | 818 | VAVAAKLLQQRLSVSK | HHHHHHHHHHHCCCCE | c1d-AT-4 | multi | 0.016 | 0.013 | 0.038 | ORD | MID|HDAC4; | 0.0 |
| 32 | fp_O | P56524 | 825 | LQQRLSVSKILIVD | HHHHCCCCEEEEEC | c2-4 | uniq | 0.014 | 0.011 | 0.073 | ORD | MID|HDAC4; | 0.286 |
| 33 | fp_O | P56524 | 878 | DEVGTGPGVGFNVNMAF | CCCCCCCCCCCEEEECC | c1c-AT-4 | uniq | 0.029 | 0.035 | 0.337 | ORD | MID|HDAC4; | 0.125 |
| 34 | fp_O | P56524 | 886 | VGFNVNMAFTGGLDPPM | CCCEEEECCCCCCCCCC | c1cR-4 | multi-selected | 0.019 | 0.031 | 0.26 | ORD | MID|HDAC4; | 0.375 |
| 35 | fp_O | P56524 | 890 | VNMAFTGGLDPPMGDAE | EEECCCCCCCCCCCCHH | c1cR-4 | multi-selected | 0.019 | 0.03 | 0.236 | ORD | MID|HDAC4; | 0.0 |
| 36 | fp_O | P56524 | 904 | DAEYLAAFRTVVMPI | CHHHHHHHHHHHCCC | c1b-4 | multi-selected | 0.01 | 0.01 | 0.12 | ORD | MID|HDAC4; | 0.0 |
| 37 | fp_O | P56524 | 905 | AEYLAAFRTVVMPI | HHHHHHHHHHHCCC | c2-AT-4 | multi-selected | 0.01 | 0.009 | 0.116 | ORD | MID|HDAC4; | 0.0 |
| 38 | fp_O | P56524 | 907 | YLAAFRTVVMPIAS | HHHHHHHHHCCCCC | c2-AT-5 | multi-selected | 0.01 | 0.009 | 0.096 | ORD | MID|HDAC4; | 0.0 |
| 39 | fp_O | P56524 | 925 | DVVLVSSGFDAVEG | CEEEECCCCCCCCC | c3-4 | uniq | 0.014 | 0.027 | 0.228 | ORD | MID|HDAC4; | 0.286 |
| 40 | fp_O | P56524 | 949 | SARCFGYLTKQLMGLAG | CCCHHHHHHHHHHHHCC | c1c-AT-4 | multi | 0.016 | 0.013 | 0.083 | ORD | MID|HDAC4; | 0.0 |
| 41 | fp_O | P56524 | 952 | CFGYLTKQLMGLAG | HHHHHHHHHHHHCC | c3-4 | multi-selected | 0.017 | 0.012 | 0.07 | ORD | MID|HDAC4; | 0.0 |
| 42 | fp_D | P56524 | 956 | LTKQLMGLAGGRIVLAL | HHHHHHHHCCCCEEEEE | c1c-AT-4 | multi | 0.016 | 0.012 | 0.123 | boundary | MID|HDAC4; | 0.0 |
| 43 | fp_D | P56524 | 957 | TKQLMGLAGGRIVLAL | HHHHHHHCCCCEEEEE | c1d-AT-4 | multi | 0.016 | 0.012 | 0.127 | boundary | MID|HDAC4; | 0.0 |
| 44 | fp_D | P56524 | 1009 | PNANAVRSMEKVMEIHS | CCHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.046 | 0.05 | 0.391 | boundary | MID|HDAC4; | 0.0 |
| 45 | fp_D | P56524 | 1010 | NANAVRSMEKVMEIHS | CHHHHHHHHHHHHHHH | c1d-4 | multi | 0.037 | 0.048 | 0.378 | boundary | MID|HDAC4; | 0.0 |
| 46 | cand_D | P56524 | 1051 | EEAETVTAMASLSVGV +++++*++*++*+*+* | CHHHHHHHCCCCCCCC | c1a-AT-4 | multi-selected | 0.969 | 0.868 | 0.536 | DISO | boundary|HDAC4; | 0.0 |
| 47 | cand_D | P56524 | 1052 | EAETVTAMASLSVGVKP ++++*++*++*+*+*++ | HHHHHHHCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.966 | 0.876 | 0.558 | DISO | boundary|HDAC4; | 0.0 |
| 48 | cand_D | P56524 | 1052 | EAETVTAMASLSVGV ++++*++*++*+*+* | HHHHHHHCCCCCCCC | c1b-4 | multi | 0.968 | 0.868 | 0.538 | DISO | boundary|HDAC4; | 0.0 |
| 49 | cand_D | P56524 | 1053 | AETVTAMASLSVGV +++*++*++*+*+* | HHHHHHCCCCCCCC | c2-AT-4 | multi | 0.967 | 0.868 | 0.545 | DISO | boundary|HDAC4; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment