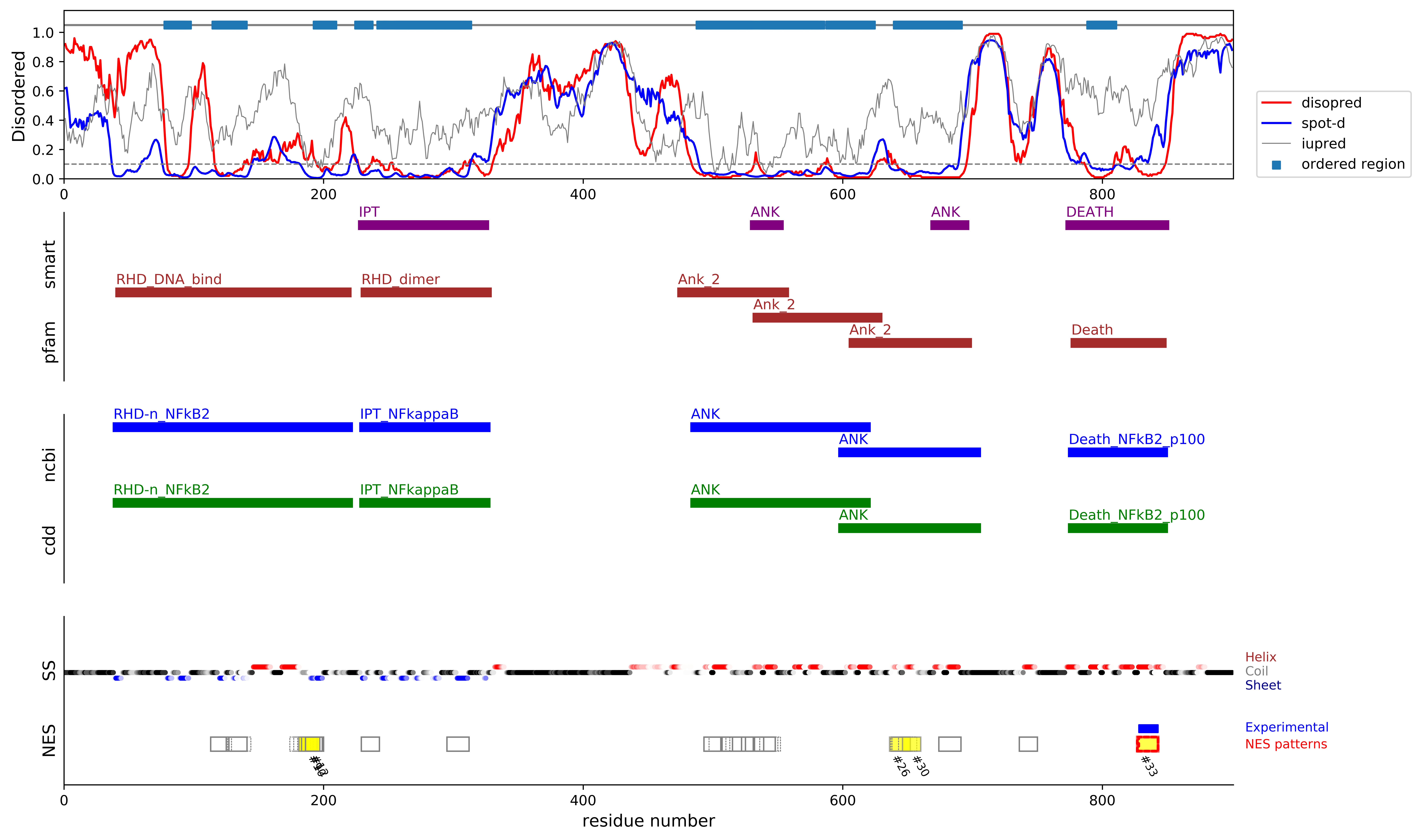

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q00653 | 113 | QCSELGICAVSVGP | CCCCCCEEEEEECC | c2-4 | uniq | 0.095 | 0.035 | 0.275 | boundary | MID|RHD-n_NFkB2; | 0.571 |
| 2 | fp_beta_D | Q00653 | 125 | GPKDMTAQFNNLGVLH | CCCCCEEEECCCCEEE | c1a-4 | multi-selected | 0.053 | 0.024 | 0.376 | boundary | MID|RHD-n_NFkB2; | 0.571 |
| 3 | fp_beta_D | Q00653 | 126 | PKDMTAQFNNLGVLH | CCCCEEEECCCCEEE | c1b-AT-4 | multi | 0.055 | 0.024 | 0.377 | boundary | MID|RHD-n_NFkB2; | 0.571 |
| 4 | fp_beta_D | Q00653 | 127 | KDMTAQFNNLGVLH | CCCEEEECCCCEEE | c2-AT-4 | multi | 0.058 | 0.022 | 0.383 | boundary | MID|RHD-n_NFkB2; | 0.571 |
| 5 | fp_D | Q00653 | 129 | MTAQFNNLGVLHVTK | CEEEECCCCEEEECC | c1b-4 | multi | 0.073 | 0.023 | 0.39 | boundary | MID|RHD-n_NFkB2; | 0.429 |
| 6 | fp_D | Q00653 | 174 | LEQEAKELKKVMDLSI | HHHHHHHHCCCCCCCC | c1d-AT-4 | multi | 0.211 | 0.082 | 0.334 | boundary | MID|RHD-n_NFkB2; | 0.0 |
| 7 | fp_D | Q00653 | 177 | EAKELKKVMDLSIVR | HHHHHCCCCCCCCEE | c1b-4 | multi | 0.209 | 0.065 | 0.262 | boundary | MID|RHD-n_NFkB2; | 0.0 |
| 8 | fp_D | Q00653 | 180 | ELKKVMDLSIVRLRF | HHCCCCCCCCEEEEE | c1b-5 | multi | 0.186 | 0.044 | 0.191 | boundary | MID|RHD-n_NFkB2; | 0.143 |
| 9 | fp_D | Q00653 | 181 | LKKVMDLSIVRLRF | HCCCCCCCCEEEEE | c2-4 | multi-selected | 0.177 | 0.039 | 0.177 | boundary | MID|RHD-n_NFkB2; | 0.143 |
| 10 | fp_D | Q00653 | 181 | LKKVMDLSIVRLRFSA | HCCCCCCCCEEEEEEE | c1a-4 | multi-selected | 0.166 | 0.035 | 0.169 | boundary | MID|RHD-n_NFkB2; | 0.286 |
| 11 | fp_beta_D | Q00653 | 183 | KVMDLSIVRLRFSAFL | CCCCCCCEEEEEEEEE | c1aR-5 | multi-selected | 0.142 | 0.023 | 0.139 | boundary | MID|RHD-n_NFkB2; | 0.571 |
| 12 | fp_D | Q00653 | 183 | KVMDLSIVRLRFSA | CCCCCCCEEEEEEE | c2-5 | multi-selected | 0.152 | 0.025 | 0.145 | boundary | MID|RHD-n_NFkB2; | 0.429 |
| 13 | fp_beta_D | Q00653 | 186 | DLSIVRLRFSAFLR | CCCCEEEEEEEEEE | c3-4 | multi-selected | 0.126 | 0.013 | 0.113 | boundary | MID|RHD-n_NFkB2; | 0.857 |
| 14 | fp_D | Q00653 | 229 | KISRMDKTAGSVRG | EEEECCCCCCCCCC | c3-AT | uniq | 0.089 | 0.042 | 0.464 | boundary | boundary|RHD-n_NFkB2; boundary|IPT_NFkappaB; | 0.143 |
| 15 | fp_beta_D | Q00653 | 295 | HKMKIERPVTVFLQLKR | CCCCCCCCEEEEEEEEE | c1c-4 | uniq | 0.065 | 0.038 | 0.347 | boundary | boundary|IPT_NFkappaB; | 0.5 |
| 16 | fp_D | Q00653 | 493 | HLAIIHGQTSVIEQ | CHHHHHCCHHHHHH | c3-AT | multi-selected | 0.018 | 0.036 | 0.222 | boundary | boundary|ANK; | 0.0 |
| 17 | fp_D | Q00653 | 497 | IHGQTSVIEQIVYVIH | HHCCHHHHHHHHHHHH | c1d-AT-4 | multi | 0.013 | 0.033 | 0.128 | boundary | boundary|ANK; | 0.0 |
| 18 | fp_D | Q00653 | 506 | QIVYVIHHAQDLGVVN | HHHHHHHHCCCCCCCC | c1a-AT-5 | multi-selected | 0.014 | 0.035 | 0.159 | boundary | MID|ANK; | 0.0 |
| 19 | fp_D | Q00653 | 507 | IVYVIHHAQDLGVVN | HHHHHHHCCCCCCCC | c1b-AT-5 | multi | 0.015 | 0.036 | 0.16 | boundary | MID|ANK; | 0.0 |
| 20 | fp_O | Q00653 | 510 | VIHHAQDLGVVNLTN | HHHHCCCCCCCCCCC | c1b-AT-5 | multi | 0.019 | 0.036 | 0.202 | ORD | MID|ANK; | 0.0 |
| 21 | fp_O | Q00653 | 515 | QDLGVVNLTNHLHQTP | CCCCCCCCCCCCCCCH | c1aR-4 | multi-selected | 0.031 | 0.031 | 0.25 | ORD | MID|ANK; | 0.0 |
| 22 | fp_O | Q00653 | 525 | HLHQTPLHLAVITG | CCCCCHHHHHHHCC | c3-AT | uniq | 0.075 | 0.019 | 0.229 | ORD | MID|ANK; | 0.0 |
| 23 | fp_O | Q00653 | 532 | HLAVITGQTSVVSFLL | HHHHHCCCHHHHHHHH | c1a-AT-5 | multi-selected | 0.054 | 0.018 | 0.136 | ORD | MID|ANK; | 0.0 |
| 24 | fp_O | Q00653 | 536 | ITGQTSVVSFLLRVGA | HCCCHHHHHHHHHCCC | c1d-AT-4 | multi | 0.028 | 0.024 | 0.104 | ORD | MID|ANK; | 0.0 |
| 25 | fp_O | Q00653 | 536 | ITGQTSVVSFLLRV | HCCCHHHHHHHHHC | c2-AT-4 | multi | 0.029 | 0.021 | 0.098 | ORD | MID|ANK; | 0.0 |
| 26 | fp_D | Q00653 | 636 | TALHLATEMEELGLVT | CCHHHHHHCCCHHHHH | c1a-4 | multi-selected | 0.08 | 0.058 | 0.449 | boundary | boundary|ANK; MID|ANK; | 0.0 |
| 27 | fp_D | Q00653 | 637 | ALHLATEMEELGLVT | CHHHHHHCCCHHHHH | c1b-AT-5 | multi | 0.075 | 0.056 | 0.434 | boundary | boundary|ANK; MID|ANK; | 0.0 |
| 28 | fp_D | Q00653 | 638 | LHLATEMEELGLVT | HHHHHHCCCHHHHH | c2-AT-4 | multi | 0.069 | 0.055 | 0.418 | boundary | boundary|ANK; MID|ANK; | 0.0 |
| 29 | fp_D | Q00653 | 643 | EMEELGLVTHLVTK | HCCCHHHHHHHHHH | c3-AT | multi | 0.037 | 0.045 | 0.355 | boundary | boundary|ANK; MID|ANK; | 0.0 |
| 30 | fp_D | Q00653 | 646 | ELGLVTHLVTKLRA | CHHHHHHHHHHHCC | c3-4 | multi-selected | 0.027 | 0.042 | 0.337 | boundary | boundary|ANK; MID|ANK; | 0.0 |
| 31 | fp_D | Q00653 | 674 | LAAGLGYPTLTRLLLKA | HHHCCCCHHHHHHHHHC | c1c-AT-4 | uniq | 0.01 | 0.074 | 0.346 | boundary | boundary|ANK; | 0.0 |
| 32 | fp_D | Q00653 | 736 | LTCSTKVKTLLLNA | CCCCHHHHHHHHHC | c2-AT-4 | uniq | 0.306 | 0.347 | 0.412 | DISO | boundary|ANK; | 0.0 |
| 33 | cand_D | Q00653 | 827 | DLAGLLEALSDMGLEE ** * * | CHHHHHHHHHHCCCHH | c1a-5 | multi-selected | 0.034 | 0.189 | 0.482 | DISO | boundary|Death_NFkB2_p100; | 0.0 |
| 34 | cand_D | Q00653 | 827 | DLAGLLEALSDMGLEE ** * * | CHHHHHHHHHHCCCHH | c1d-AT-5 | multi | 0.034 | 0.189 | 0.482 | DISO | boundary|Death_NFkB2_p100; | 0.0 |
| 35 | cand_D | Q00653 | 828 | LAGLLEALSDMGLEE ** * * | HHHHHHHHHHCCCHH | c1b-4 | multi | 0.032 | 0.192 | 0.488 | DISO | boundary|Death_NFkB2_p100; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment