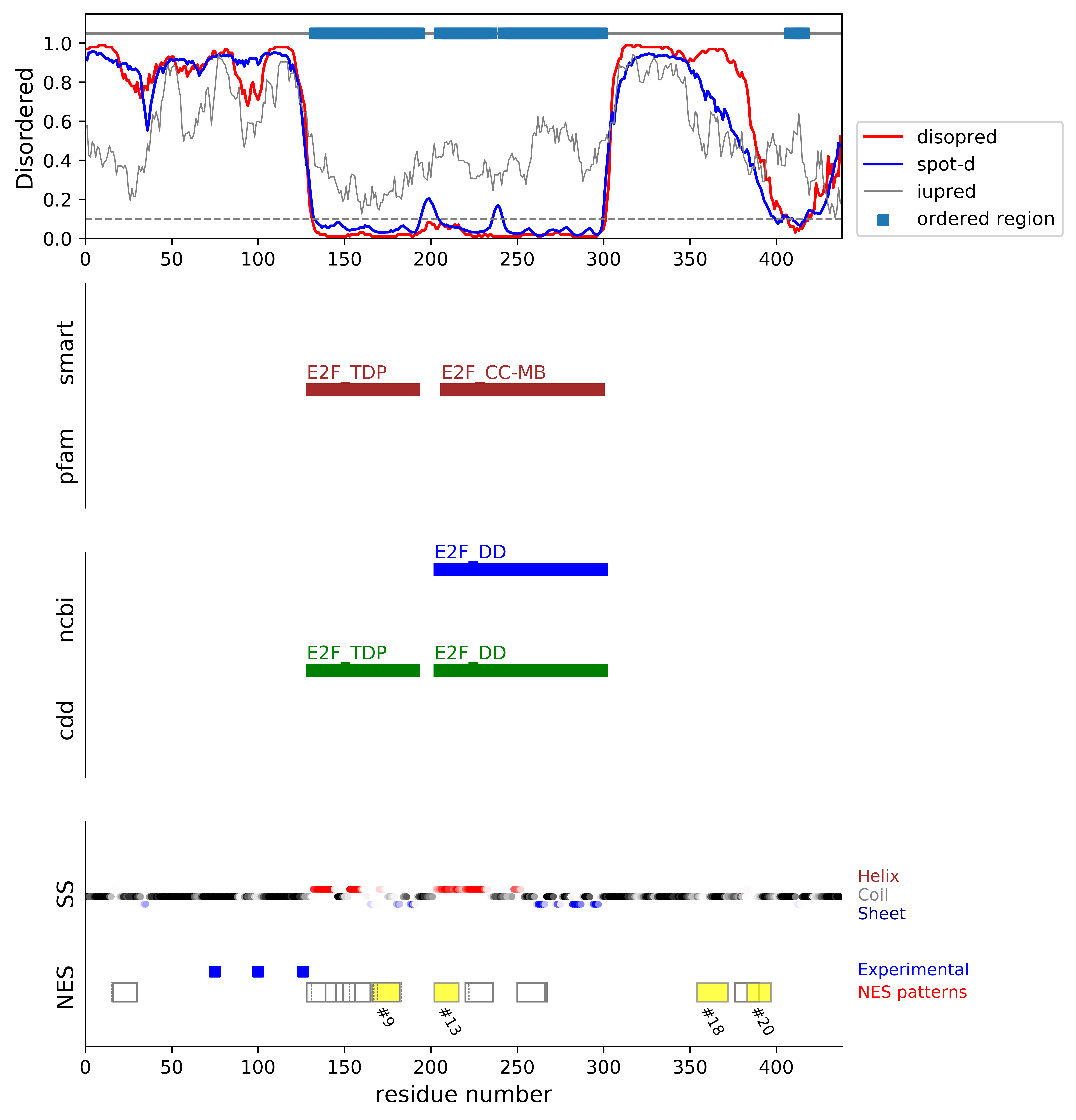

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q01094 | 15 | ALEALLGAGALRLLD | CHHHCCCCCCCCCCC | c1b-AT-5 | multi | 0.861 | 0.888 | 0.329 | DISO | 0.0 | |
| 2 | fp_D | Q01094 | 16 | LEALLGAGALRLLD | HHHCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.853 | 0.886 | 0.321 | DISO | 0.0 | |
| 3 | fp_D | Q01094 | 128 | YETSLNLTTKRFLELLS | CCCCHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.08 | 0.122 | 0.406 | boundary | boundary|E2F_TDP; | 0.0 |
| 4 | fp_D | Q01094 | 131 | SLNLTTKRFLELLS | CHHHHHHHHHHHHH | c3-AT | multi | 0.029 | 0.077 | 0.372 | boundary | boundary|E2F_TDP; | 0.0 |
| 5 | fp_D | Q01094 | 139 | FLELLSHSADGVVD | HHHHHHHCCCCCCC | c3-AT | multi | 0.011 | 0.063 | 0.28 | boundary | boundary|E2F_TDP; | 0.0 |
| 6 | fp_D | Q01094 | 139 | FLELLSHSADGVVDLNW | HHHHHHHCCCCCCCHHH | c1c-AT-5 | multi-selected | 0.012 | 0.06 | 0.285 | boundary | boundary|E2F_TDP; | 0.0 |
| 7 | fp_O | Q01094 | 149 | GVVDLNWAAEVLKVQK | CCCCHHHHHHHHHHCE | c1a-AT-5 | multi-selected | 0.019 | 0.052 | 0.224 | ORD | MID|E2F_TDP; | 0.0 |
| 8 | fp_O | Q01094 | 149 | GVVDLNWAAEVLKVQK | CCCCHHHHHHHHHHCE | c1d-AT-5 | multi | 0.019 | 0.052 | 0.224 | ORD | MID|E2F_TDP; | 0.0 |
| 9 | fp_D | Q01094 | 166 | RIYDITNVLEGIQLIA | EEEHHHHHHCCCCCEE | c1a-5 | multi-selected | 0.011 | 0.036 | 0.22 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 10 | fp_D | Q01094 | 166 | RIYDITNVLEGIQLIA | EEEHHHHHHCCCCCEE | c1d-5 | multi | 0.011 | 0.036 | 0.22 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 11 | fp_D | Q01094 | 167 | IYDITNVLEGIQLIA | EEHHHHHHCCCCCEE | c1b-AT-4 | multi | 0.011 | 0.035 | 0.217 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 12 | fp_D | Q01094 | 169 | DITNVLEGIQLIAK | HHHHHHCCCCCEEE | c3-4 | multi | 0.011 | 0.036 | 0.216 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 13 | fp_D | Q01094 | 202 | RLEGLTQDLRQLQE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.062 | 0.08 | 0.459 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 14 | fp_D | Q01094 | 220 | LDHLMNICTTQLRLLS | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi-selected | 0.016 | 0.04 | 0.341 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 15 | fp_D | Q01094 | 222 | HLMNICTTQLRLLS | HHHHHHHHHHHHCC | c2-AT-5 | multi | 0.015 | 0.04 | 0.334 | boundary | boundary|E2F_TDP; boundary|E2F_DD; | 0.0 |
| 16 | fp_D | Q01094 | 250 | DLRSIADPAEQMVMVIK | HHHCCCCCCCCEEEEEE | c1c-AT-5 | multi-selected | 0.016 | 0.028 | 0.45 | __ | MID|E2F_DD; | 0.125 |
| 17 | fp_D | Q01094 | 250 | DLRSIADPAEQMVMVI | HHHCCCCCCCCEEEEE | c1a-AT-5 | multi-selected | 0.016 | 0.028 | 0.442 | __ | MID|E2F_DD; | 0.0 |
| 18 | fp_D | Q01094 | 354 | EQEPLLSRMGSLRAPVDE | CCCCCCCCCCCCCCCCCC | c4-4 | uniq | 0.959 | 0.716 | 0.532 | DISO | 0.0 | |
| 19 | fp_D | Q01094 | 376 | PLVAADSLLEHVRE | CCCCCCCHHHHCCC | c3-AT | uniq | 0.713 | 0.428 | 0.419 | DISO | 0.0 | |
| 20 | fp_D | Q01094 | 383 | LLEHVREDFSGLLP | HHHHCCCCCCCCCC | c3-4 | uniq | 0.471 | 0.289 | 0.377 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment