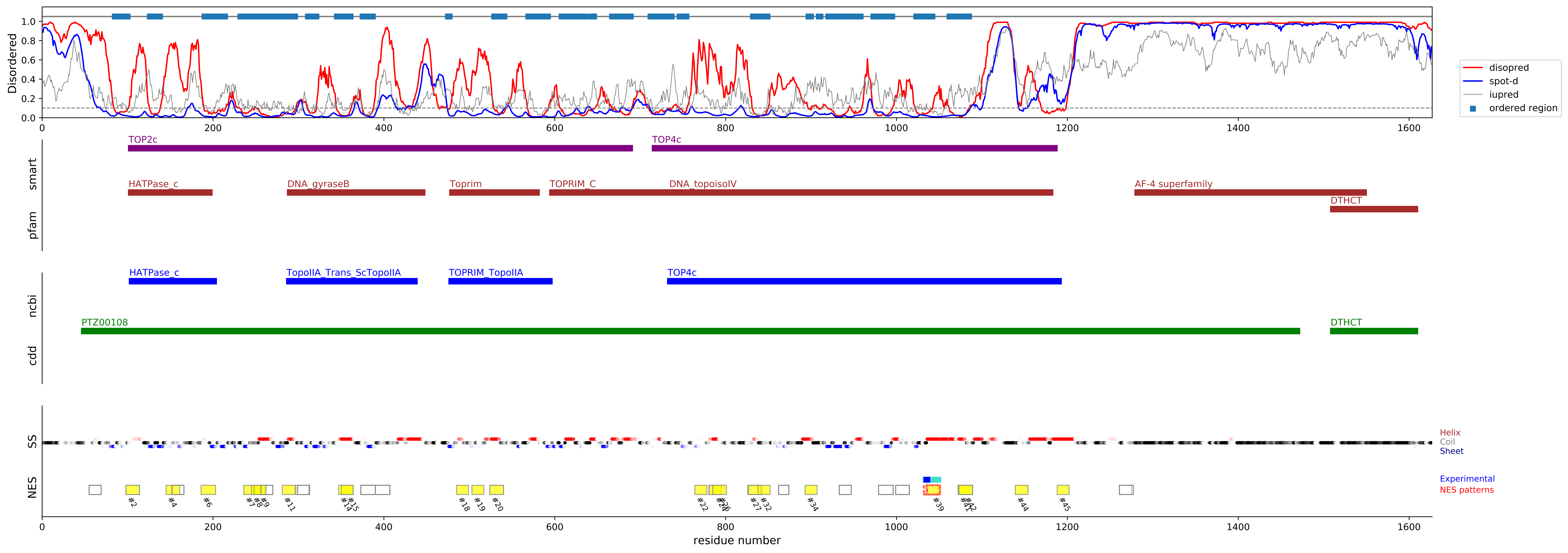

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q02880 | 55 | YQKKTQLEHILLRP | HHHCCCCHHHHCCC | c2-AT-4 | uniq | 0.846 | 0.196 | 0.248 | DISO | boundary|PTZ00108; | 0.0 |
| 2 | fp_D | Q02880 | 98 | FVPGLYKIFDEILVNA | CCCCCCCCHHHHHHHH | c1a-5 | multi-selected | 0.355 | 0.013 | 0.123 | boundary | MID|PTZ00108; | 0.0 |
| 3 | fp_D | Q02880 | 98 | FVPGLYKIFDEILVNA | CCCCCCCCHHHHHHHH | c1d-5 | multi | 0.355 | 0.013 | 0.123 | boundary | MID|PTZ00108; | 0.0 |
| 4 | fp_D | Q02880 | 145 | GIPVVEHKVEKVYVPA | CCCEEECCCCCCCCCE | c1a-5 | uniq | 0.688 | 0.016 | 0.132 | boundary | MID|PTZ00108; | 0.286 |
| 5 | fp_D | Q02880 | 152 | KVEKVYVPALIFGQ | CCCCCCCCEEEEEE | c3-AT | uniq | 0.594 | 0.013 | 0.082 | boundary | MID|PTZ00108; | 0.286 |
| 6 | fp_D | Q02880 | 186 | YGAKLCNIFSTKFTVET | CCCHHHHHCCCEEEEEE | c1c-4 | uniq | 0.096 | 0.014 | 0.104 | boundary | MID|PTZ00108; | 0.125 |
| 7 | fp_D | Q02880 | 236 | TCITFQPDLSKFKMEK | EEEEEEECCCCCCCCC | c1a-4 | uniq | 0.072 | 0.026 | 0.121 | boundary | MID|PTZ00108; | 0.429 |
| 8 | fp_D | Q02880 | 245 | SKFKMEKLDKDIVALM | CCCCCCCCCHHHHHHH | c1aR-5 | multi-selected | 0.072 | 0.029 | 0.129 | __ | MID|PTZ00108; | 0.0 |
| 9 | fp_D | Q02880 | 248 | KMEKLDKDIVALMT | CCCCCCHHHHHHHH | c3-4 | multi-selected | 0.065 | 0.026 | 0.131 | boundary | MID|PTZ00108; | 0.0 |
| 10 | fp_O | Q02880 | 256 | IVALMTRRAYDLAG | HHHHHHHHHHHHHC | c3-AT | uniq | 0.024 | 0.01 | 0.111 | ORD | MID|PTZ00108; | 0.0 |
| 11 | fp_D | Q02880 | 281 | KKLPVNGFRSYVDLYV | EECCCCCHHHHHHHHH | c1d-4 | uniq | 0.046 | 0.043 | 0.085 | boundary | MID|PTZ00108; | 0.0 |
| 12 | fp_D | Q02880 | 296 | VKDKLDETGVALKVIH | HCCCCCCCCCCCEEEE | c1d-AT-4 | multi | 0.147 | 0.094 | 0.118 | boundary | MID|PTZ00108; | 0.0 |
| 13 | fp_D | Q02880 | 299 | KLDETGVALKVIHE | CCCCCCCCCEEEEE | c3-AT | multi-selected | 0.151 | 0.094 | 0.125 | boundary | MID|PTZ00108; | 0.143 |
| 14 | fp_D | Q02880 | 347 | VDYVVDQVVGKLIEVVK | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.045 | 0.021 | 0.117 | boundary | MID|PTZ00108; | 0.0 |
| 15 | fp_D | Q02880 | 350 | VVDQVVGKLIEVVK | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.045 | 0.021 | 0.108 | boundary | MID|PTZ00108; | 0.0 |
| 16 | fp_beta_D | Q02880 | 373 | KPFQVKNHIWVFINCLI | CCHHHCCCEEEEEEEEE | c1cR-5 | uniq | 0.066 | 0.021 | 0.065 | boundary | MID|PTZ00108; | 0.5 |
| 17 | fp_D | Q02880 | 390 | ENPTFDSQTKENMTLQP | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.696 | 0.172 | 0.317 | boundary | MID|PTZ00108; | 0.0 |
| 18 | fp_D | Q02880 | 485 | SAKSLAVSGLGVIG | CHHHHHHCCCCCCC | c2-4 | uniq | 0.477 | 0.063 | 0.151 | DISO | MID|PTZ00108; | 0.0 |
| 19 | fp_D | Q02880 | 503 | GVFPLRGKILNVRE | EECCCCCCCCEECC | c3-4 | uniq | 0.568 | 0.034 | 0.169 | boundary | MID|PTZ00108; | 0.0 |
| 20 | fp_D | Q02880 | 524 | ENAEINNIIKIVGLQY | HHHHHHHHHHHHCCCC | c1a-4 | multi-selected | 0.131 | 0.044 | 0.146 | boundary | MID|PTZ00108; | 0.0 |
| 21 | fp_D | Q02880 | 524 | ENAEINNIIKIVGLQY | HHHHHHHHHHHHCCCC | c1d-4 | multi | 0.131 | 0.044 | 0.146 | boundary | MID|PTZ00108; | 0.0 |
| 22 | fp_D | Q02880 | 764 | KVAQLAGSVAEMSA | EEEEEHHHHHHHCC | c3-4 | uniq | 0.593 | 0.056 | 0.267 | boundary | MID|PTZ00108; | 0.286 |
| 23 | fp_D | Q02880 | 780 | HGEQALMMTIVNLAQ | CHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.586 | 0.03 | 0.143 | DISO | MID|PTZ00108; | 0.0 |
| 24 | fp_D | Q02880 | 781 | GEQALMMTIVNLAQ | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.589 | 0.028 | 0.141 | DISO | MID|PTZ00108; | 0.0 |
| 25 | fp_D | Q02880 | 785 | LMMTIVNLAQNFVG | HHHHHHHHHHCCCC | c3-AT | multi | 0.479 | 0.026 | 0.119 | DISO | MID|PTZ00108; | 0.0 |
| 26 | fp_D | Q02880 | 785 | LMMTIVNLAQNFVGSN | HHHHHHHHHHCCCCCC | c1aR-4 | multi-selected | 0.451 | 0.027 | 0.12 | DISO | MID|PTZ00108; | 0.0 |
| 27 | fp_D | Q02880 | 826 | YIFTMLSTLARLLFPA | CCCCCCHHHHHCCCCC | c1a-5 | multi-selected | 0.122 | 0.014 | 0.058 | boundary | MID|PTZ00108; | 0.0 |
| 28 | fp_D | Q02880 | 826 | YIFTMLSTLARLLFPA | CCCCCCHHHHHCCCCC | c1d-AT-5 | multi | 0.122 | 0.014 | 0.058 | boundary | MID|PTZ00108; | 0.0 |
| 29 | fp_D | Q02880 | 826 | YIFTMLSTLARLLFPAV | CCCCCCHHHHHCCCCCC | c1cR-4 | multi | 0.116 | 0.015 | 0.056 | boundary | MID|PTZ00108; | 0.0 |
| 30 | fp_D | Q02880 | 827 | IFTMLSTLARLLFPA | CCCCCHHHHHCCCCC | c1b-5 | multi | 0.103 | 0.014 | 0.047 | boundary | MID|PTZ00108; | 0.0 |
| 31 | fp_D | Q02880 | 827 | IFTMLSTLARLLFP | CCCCCHHHHHCCCC | c3-AT | multi | 0.109 | 0.014 | 0.049 | boundary | MID|PTZ00108; | 0.0 |
| 32 | fp_D | Q02880 | 838 | LFPAVDDNLLKFLY | CCCCCCCCCCCCCC | c3-4 | uniq | 0.044 | 0.026 | 0.083 | boundary | MID|PTZ00108; | 0.0 |
| 33 | fp_beta_D | Q02880 | 862 | YIPIIPMVLING | ECCCCCEEEECC | c2-rev | uniq | 0.354 | 0.013 | 0.074 | boundary | MID|PTZ00108; | 0.5 |
| 34 | fp_D | Q02880 | 893 | IVNNVRRMLDGLDP | HHHHHHHHHCCCCC | c3-4 | uniq | 0.119 | 0.03 | 0.321 | boundary | MID|PTZ00108; | 0.0 |
| 35 | fp_beta_D | Q02880 | 933 | EIFVVDRNTVEITE | EEEEEECCEEEEEE | c3-AT | uniq | 0.051 | 0.012 | 0.165 | boundary | MID|PTZ00108; | 0.714 |
| 36 | fp_beta_D | Q02880 | 979 | KEYHTDTTVKFVVKMTE | CCCCCCCEEEEEEEECH | c1c-AT-4 | uniq | 0.037 | 0.037 | 0.224 | boundary | MID|PTZ00108; | 0.625 |
| 37 | fp_D | Q02880 | 999 | AQAEAAGLHKVFKLQT | HHHHHHCHHHHHCCCC | c1d-AT-4 | uniq | 0.298 | 0.045 | 0.172 | boundary | MID|PTZ00108; | 0.0 |
| 38 | cand_D | Q02880 | 1032 | KYETVQDILKEFFDLRL +**++++++++++++ | CCCCHHHHHHHHHHHHH | c1c-4 | multi | 0.114 | 0.014 | 0.075 | boundary | MID|PTZ00108; | 0.0 |
| 39 | cand_D | Q02880 | 1035 | TVQDILKEFFDLRLSY **+++++++++++++ | CHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.151 | 0.012 | 0.067 | boundary | MID|PTZ00108; | 0.0 |
| 40 | cand_D | Q02880 | 1036 | VQDILKEFFDLRLSY *+++++++++++++ | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.159 | 0.011 | 0.061 | boundary | MID|PTZ00108; | 0.0 |
| 41 | fp_D | Q02880 | 1072 | QARFILEKIQGKITIEN | HHHHHHHHHCCCEEECC | c1c-4 | multi-selected | 0.052 | 0.048 | 0.265 | boundary | MID|PTZ00108; | 0.0 |
| 42 | fp_D | Q02880 | 1073 | ARFILEKIQGKITIEN | HHHHHHHHCCCEEECC | c1aR-4 | multi-selected | 0.054 | 0.049 | 0.264 | boundary | MID|PTZ00108; | 0.0 |
| 43 | fp_D | Q02880 | 1073 | ARFILEKIQGKITIEN | HHHHHHHHCCCEEECC | c1d-4 | multi | 0.054 | 0.049 | 0.264 | boundary | MID|PTZ00108; | 0.0 |
| 44 | fp_D | Q02880 | 1139 | SGPDFNYILNMSLWS | CCCCCCHHCCCCCCC | c1b-4 | uniq | 0.274 | 0.146 | 0.353 | DISO | MID|PTZ00108; | 0.0 |
| 45 | fp_D | Q02880 | 1188 | DLAAFVEELDKVES | HHHHHHHHHHHHHH | c3-4 | uniq | 0.109 | 0.196 | 0.431 | DISO | MID|PTZ00108; | 0.0 |
| 46 | fp_D | Q02880 | 1261 | KKGDLDTAAVKVEFDE | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.987 | 0.961 | 0.479 | DISO | MID|PTZ00108; | 0.0 |
| 47 | fp_D | Q02880 | 1261 | KKGDLDTAAVKVEFDE | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.987 | 0.961 | 0.479 | DISO | MID|PTZ00108; | 0.0 |
| 48 | fp_D | Q02880 | 1261 | KKGDLDTAAVKVEF | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.988 | 0.962 | 0.475 | DISO | MID|PTZ00108; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment