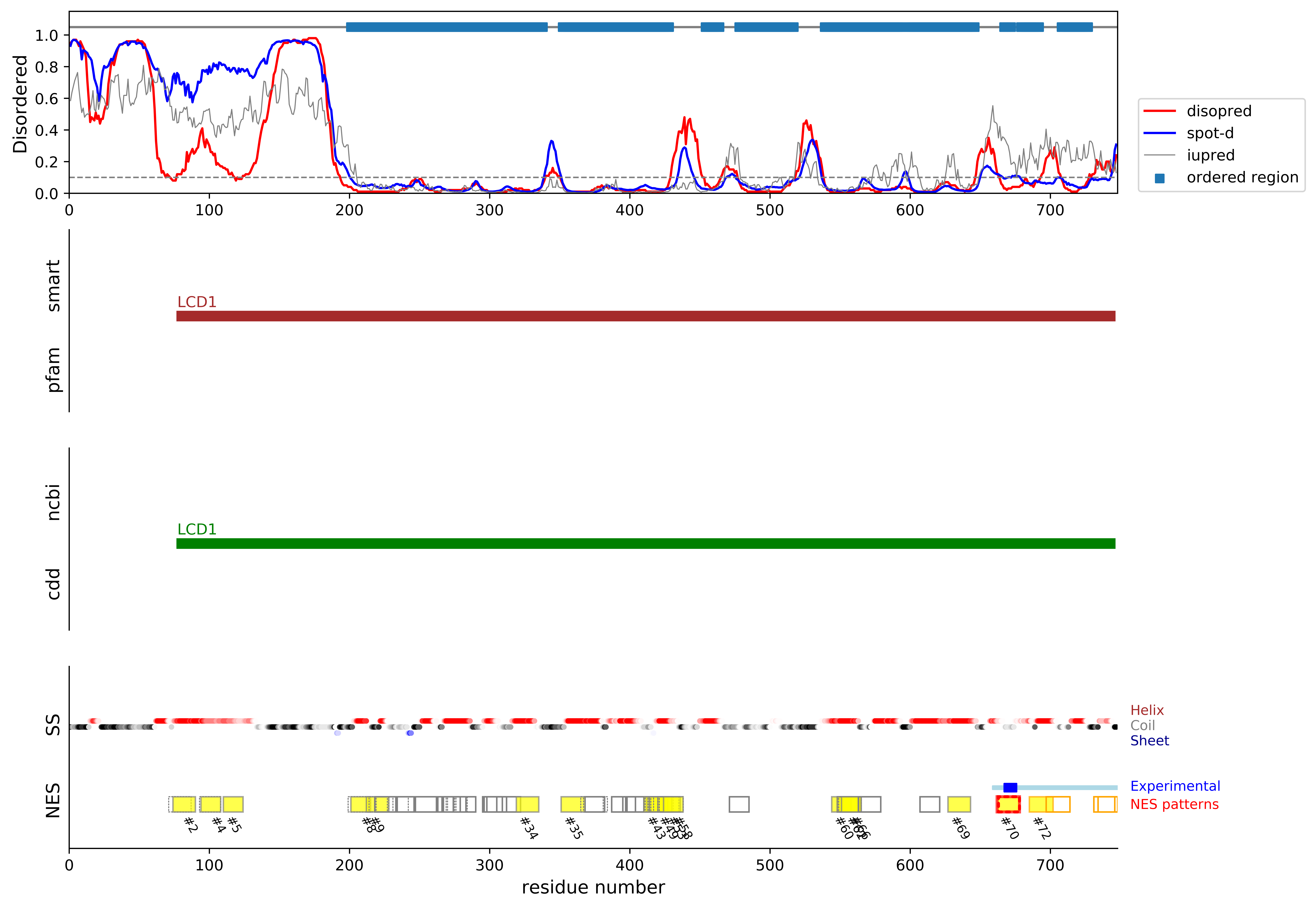

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q04377 | 71 | AQGEASMLRDKINFLN | HHCHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.135 | 0.684 | 0.512 | DISO | boundary|LCD1; | 0.0 |
| 2 | fp_D | Q04377 | 74 | EASMLRDKINFLNIER | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.166 | 0.68 | 0.488 | DISO | boundary|LCD1; | 0.0 |
| 3 | fp_D | Q04377 | 93 | KNIQAVKVNELQVKH | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.286 | 0.777 | 0.457 | DISO | boundary|LCD1; | 0.0 |
| 4 | fp_D | Q04377 | 94 | NIQAVKVNELQVKH | HHHHHHHHHHHHHH | c2-5 | multi-selected | 0.281 | 0.782 | 0.458 | DISO | boundary|LCD1; | 0.0 |
| 5 | fp_D | Q04377 | 110 | ELAKLKQELQKLED | HHHHHHHHHHHHHH | c3-4 | uniq | 0.118 | 0.785 | 0.438 | DISO | MID|LCD1; | 0.0 |
| 6 | fp_D | Q04377 | 199 | IPDETSLFLESILLHQ | CCCCHHHHHHHHHHCC | c1a-AT-4 | multi | 0.019 | 0.064 | 0.103 | boundary | MID|LCD1; | 0.0 |
| 7 | fp_D | Q04377 | 199 | IPDETSLFLESILLHQ | CCCCHHHHHHHHHHCC | c1d-AT-4 | multi | 0.019 | 0.064 | 0.103 | boundary | MID|LCD1; | 0.0 |
| 8 | fp_D | Q04377 | 201 | DETSLFLESILLHQ | CCHHHHHHHHHHCC | c2-4 | multi-selected | 0.016 | 0.056 | 0.081 | boundary | MID|LCD1; | 0.0 |
| 9 | fp_D | Q04377 | 212 | LHQIIGADLSTIEILN | HCCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.01 | 0.042 | 0.038 | boundary | MID|LCD1; | 0.0 |
| 10 | fp_D | Q04377 | 214 | QIIGADLSTIEILN | CCCCCCCCHHHHHH | c2-AT-5 | multi | 0.01 | 0.041 | 0.037 | __ | MID|LCD1; | 0.0 |
| 11 | fp_O | Q04377 | 218 | ADLSTIEILNRLKLDY | CCCCHHHHHHCCCCCC | c1a-AT-4 | multi-selected | 0.01 | 0.04 | 0.037 | ORD | MID|LCD1; | 0.0 |
| 12 | fp_O | Q04377 | 218 | ADLSTIEILNRLKLDY | CCCCHHHHHHCCCCCC | c1d-AT-4 | multi | 0.01 | 0.04 | 0.037 | ORD | MID|LCD1; | 0.0 |
| 13 | fp_O | Q04377 | 219 | DLSTIEILNRLKLDY | CCCHHHHHHCCCCCC | c1b-5 | multi | 0.01 | 0.039 | 0.035 | ORD | MID|LCD1; | 0.0 |
| 14 | fp_O | Q04377 | 227 | NRLKLDYITEFKFKN | HCCCCCCCCCCCCCC | c1b-4 | multi | 0.016 | 0.047 | 0.025 | ORD | MID|LCD1; | 0.0 |
| 15 | fp_O | Q04377 | 231 | LDYITEFKFKNFVIAK | CCCCCCCCCCCEEECC | c1a-AT-4 | multi | 0.033 | 0.05 | 0.027 | ORD | MID|LCD1; | 0.0 |
| 16 | fp_O | Q04377 | 233 | YITEFKFKNFVIAK | CCCCCCCCCEEECC | c2-5 | multi-selected | 0.036 | 0.05 | 0.026 | ORD | MID|LCD1; | 0.143 |
| 17 | fp_O | Q04377 | 246 | KGAPIGKSIVSLLLRC | CCCCCHHHHHHHHHHH | c1a-4 | uniq | 0.053 | 0.048 | 0.029 | ORD | MID|LCD1; | 0.0 |
| 18 | fp_O | Q04377 | 263 | KTLTLDRFIDTLLEDI | CCCCHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.018 | 0.022 | 0.013 | ORD | MID|LCD1; | 0.0 |
| 19 | fp_O | Q04377 | 266 | TLDRFIDTLLEDIAVLI | CHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.021 | 0.017 | 0.016 | ORD | MID|LCD1; | 0.0 |
| 20 | fp_O | Q04377 | 267 | LDRFIDTLLEDIAVLI | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.021 | 0.016 | 0.016 | ORD | MID|LCD1; | 0.0 |
| 21 | fp_O | Q04377 | 267 | LDRFIDTLLEDIAVLI | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.021 | 0.016 | 0.016 | ORD | MID|LCD1; | 0.0 |
| 22 | fp_O | Q04377 | 269 | RFIDTLLEDIAVLI | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.021 | 0.014 | 0.016 | ORD | MID|LCD1; | 0.0 |
| 23 | fp_O | Q04377 | 270 | FIDTLLEDIAVLIK | HHHHHHHHHHHHHH | c3-4 | multi | 0.021 | 0.014 | 0.017 | ORD | MID|LCD1; | 0.0 |
| 24 | fp_O | Q04377 | 274 | LLEDIAVLIKEISVHP | HHHHHHHHHHHHHCCC | c1a-5 | multi-selected | 0.029 | 0.022 | 0.022 | ORD | MID|LCD1; | 0.0 |
| 25 | fp_O | Q04377 | 274 | LLEDIAVLIKEISVHP | HHHHHHHHHHHHHCCC | c1d-5 | multi | 0.029 | 0.022 | 0.022 | ORD | MID|LCD1; | 0.0 |
| 26 | fp_O | Q04377 | 275 | LEDIAVLIKEISVHP | HHHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.029 | 0.023 | 0.023 | ORD | MID|LCD1; | 0.0 |
| 27 | fp_O | Q04377 | 276 | EDIAVLIKEISVHP | HHHHHHHHHHHCCC | c2-4 | multi | 0.03 | 0.023 | 0.024 | ORD | MID|LCD1; | 0.0 |
| 28 | fp_O | Q04377 | 295 | AVPFLVALMYQIVQFRP | CHHHHHHHHHHHCCCCC | c1c-5 | multi-selected | 0.013 | 0.016 | 0.01 | ORD | MID|LCD1; | 0.0 |
| 29 | fp_O | Q04377 | 295 | AVPFLVALMYQIVQ | CHHHHHHHHHHHCC | c3-4 | multi-selected | 0.014 | 0.013 | 0.009 | ORD | MID|LCD1; | 0.0 |
| 30 | fp_O | Q04377 | 296 | VPFLVALMYQIVQFRP | HHHHHHHHHHHCCCCC | c1aR-4 | multi-selected | 0.012 | 0.016 | 0.01 | ORD | MID|LCD1; | 0.0 |
| 31 | fp_O | Q04377 | 296 | VPFLVALMYQIVQFRP | HHHHHHHHHHHCCCCC | c1d-4 | multi | 0.012 | 0.016 | 0.01 | ORD | MID|LCD1; | 0.0 |
| 32 | fp_O | Q04377 | 298 | FLVALMYQIVQFRP | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.011 | 0.016 | 0.01 | ORD | MID|LCD1; | 0.0 |
| 33 | fp_O | Q04377 | 305 | QIVQFRPSATHNLALKD | HHCCCCCCCCCHHHHHH | c1c-AT-5 | uniq | 0.017 | 0.025 | 0.011 | ORD | MID|LCD1; | 0.0 |
| 34 | fp_D | Q04377 | 319 | LKDCFLFICDLIRIYH | HHHHHHHHHHHHHHCC | c1d-4 | uniq | 0.021 | 0.013 | 0.004 | boundary | MID|LCD1; | 0.0 |
| 35 | fp_D | Q04377 | 351 | EPQIFQYELIDYLIISY | CCCHHHHHHHHHHHHHH | c1c-4 | uniq | 0.021 | 0.022 | 0.009 | boundary | MID|LCD1; | 0.0 |
| 36 | fp_D | Q04377 | 365 | ISYSFDLLEGILRVLQ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.022 | 0.009 | 0.013 | __ | MID|LCD1; | 0.0 |
| 37 | fp_O | Q04377 | 367 | YSFDLLEGILRVLQSHP | HHHHHHHHHHHHHHCCC | c1cR-4 | multi | 0.026 | 0.015 | 0.017 | ORD | MID|LCD1; | 0.0 |
| 38 | fp_O | Q04377 | 368 | SFDLLEGILRVLQS | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.026 | 0.012 | 0.015 | ORD | MID|LCD1; | 0.0 |
| 39 | fp_O | Q04377 | 387 | YMEFFDENILKSFEFVY | HHHHHHHHHHHHHHHHH | c1c-5 | uniq | 0.017 | 0.025 | 0.023 | ORD | MID|LCD1; | 0.0 |
| 40 | fp_O | Q04377 | 395 | ILKSFEFVYKLALTI | HHHHHHHHHHHHHHC | c1b-5 | multi-selected | 0.014 | 0.027 | 0.007 | ORD | MID|LCD1; | 0.0 |
| 41 | fp_D | Q04377 | 397 | KSFEFVYKLALTISYKP | HHHHHHHHHHHHCCCCC | c1cR-4 | multi-selected | 0.015 | 0.032 | 0.007 | boundary | MID|LCD1; | 0.0 |
| 42 | fp_O | Q04377 | 398 | SFEFVYKLALTISY | HHHHHHHHHHHCCC | c3-AT | multi-selected | 0.016 | 0.031 | 0.007 | ORD | MID|LCD1; | 0.0 |
| 43 | fp_D | Q04377 | 410 | SYKPMVNVIFSAVEVVN | CCCCCCEEHHHHHHHHH | c1c-4 | multi-selected | 0.014 | 0.033 | 0.009 | boundary | MID|LCD1; | 0.25 |
| 44 | fp_D | Q04377 | 411 | YKPMVNVIFSAVEVVN | CCCCCEEHHHHHHHHH | c1a-4 | multi | 0.013 | 0.032 | 0.009 | boundary | MID|LCD1; | 0.286 |
| 45 | fp_D | Q04377 | 411 | YKPMVNVIFSAVEVVN | CCCCCEEHHHHHHHHH | c1d-4 | multi | 0.013 | 0.032 | 0.009 | boundary | MID|LCD1; | 0.286 |
| 46 | fp_D | Q04377 | 413 | PMVNVIFSAVEVVNIIT | CCCEEHHHHHHHHHHHH | c1c-AT-5 | multi | 0.023 | 0.03 | 0.01 | boundary | MID|LCD1; | 0.125 |
| 47 | fp_D | Q04377 | 413 | PMVNVIFSAVEVVN | CCCEEHHHHHHHHH | c2-5 | multi | 0.013 | 0.029 | 0.009 | boundary | MID|LCD1; | 0.286 |
| 48 | fp_D | Q04377 | 413 | PMVNVIFSAVEVVN | CCCEEHHHHHHHHH | c3-AT | multi | 0.013 | 0.029 | 0.009 | boundary | MID|LCD1; | 0.286 |
| 49 | fp_D | Q04377 | 414 | MVNVIFSAVEVVNIIT | CCEEHHHHHHHHHHHH | c1a-5 | multi-selected | 0.024 | 0.029 | 0.01 | boundary | MID|LCD1; | 0.0 |
| 50 | fp_D | Q04377 | 414 | MVNVIFSAVEVVNIIT | CCEEHHHHHHHHHHHH | c1d-AT-5 | multi | 0.024 | 0.029 | 0.01 | boundary | MID|LCD1; | 0.0 |
| 51 | fp_D | Q04377 | 415 | VNVIFSAVEVVNIIT | CEEHHHHHHHHHHHH | c1b-4 | multi | 0.025 | 0.028 | 0.01 | boundary | MID|LCD1; | 0.0 |
| 52 | fp_D | Q04377 | 417 | VIFSAVEVVNIITS | EHHHHHHHHHHHHH | c3-AT | multi | 0.037 | 0.029 | 0.013 | boundary | MID|LCD1; | 0.0 |
| 53 | fp_D | Q04377 | 420 | SAVEVVNIITSIILNM | HHHHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.122 | 0.045 | 0.021 | boundary | MID|LCD1; | 0.0 |
| 54 | fp_D | Q04377 | 420 | SAVEVVNIITSIILNM | HHHHHHHHHHHHHHHC | c1d-4 | multi | 0.122 | 0.045 | 0.021 | boundary | MID|LCD1; | 0.0 |
| 55 | fp_D | Q04377 | 421 | AVEVVNIITSIILNMDN | HHHHHHHHHHHHHHCCC | c1c-AT-5 | multi | 0.16 | 0.068 | 0.023 | boundary | MID|LCD1; | 0.0 |
| 56 | fp_D | Q04377 | 421 | AVEVVNIITSIILNM | HHHHHHHHHHHHHHC | c1b-5 | multi | 0.13 | 0.046 | 0.021 | boundary | MID|LCD1; | 0.0 |
| 57 | fp_D | Q04377 | 421 | AVEVVNIITSIILN | HHHHHHHHHHHHHH | c3-AT | multi | 0.111 | 0.039 | 0.021 | boundary | MID|LCD1; | 0.0 |
| 58 | fp_D | Q04377 | 424 | VVNIITSIILNMDN | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.192 | 0.078 | 0.026 | boundary | MID|LCD1; | 0.0 |
| 59 | fp_D | Q04377 | 471 | YNENVDTTTLHMSK | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.086 | 0.082 | 0.181 | boundary | MID|LCD1; | 0.0 |
| 60 | fp_D | Q04377 | 544 | WLLKLKDEVLNIFE | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.017 | 0.02 | boundary | MID|LCD1; | 0.0 |
| 61 | fp_D | Q04377 | 548 | LKDEVLNIFENLLMIYG | HHHHHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.013 | 0.021 | 0.027 | boundary | MID|LCD1; | 0.0 |
| 62 | fp_D | Q04377 | 548 | LKDEVLNIFENLLMIY | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.012 | 0.019 | 0.025 | boundary | MID|LCD1; | 0.0 |
| 63 | fp_D | Q04377 | 548 | LKDEVLNIFENLLMIY | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.012 | 0.019 | 0.025 | boundary | MID|LCD1; | 0.0 |
| 64 | fp_D | Q04377 | 549 | KDEVLNIFENLLMIYG | HHHHHHHHHHHHHHHC | c1d-4 | multi | 0.013 | 0.021 | 0.027 | boundary | MID|LCD1; | 0.0 |
| 65 | fp_D | Q04377 | 549 | KDEVLNIFENLLMIY | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.012 | 0.019 | 0.024 | boundary | MID|LCD1; | 0.0 |
| 66 | fp_D | Q04377 | 551 | EVLNIFENLLMIYG | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.014 | 0.022 | 0.028 | boundary | MID|LCD1; | 0.0 |
| 67 | fp_O | Q04377 | 563 | YGDDATIVNGEMLIHS | HCCCCCCCCCCHHHHH | c1d-AT-4 | uniq | 0.023 | 0.056 | 0.091 | ORD | MID|LCD1; | 0.0 |
| 68 | fp_O | Q04377 | 607 | LIEHTLTIIYRLWK | HHHHHHHHHHHHHH | c3-AT | uniq | 0.015 | 0.009 | 0.04 | ORD | MID|LCD1; | 0.0 |
| 69 | fp_D | Q04377 | 627 | REEQIKQVESQLIMSL | HHHHHHHHHHHHHHHH | c1d-4 | uniq | 0.034 | 0.023 | 0.115 | boundary | MID|LCD1; | 0.0 |
| 70 | cand_D | Q04377 | 662 | DHRHLVDSLHDLTIKD ........*..*.... | HHHHHCCCCCCCCHHH | c1a-4 | multi-selected | 0.051 | 0.105 | 0.28 | boundary | MID|LCD1; | 0.0 |
| 71 | cand_D | Q04377 | 663 | HRHLVDSLHDLTIKD .......*..*.... | HHHHCCCCCCCCHHH | c1b-4 | multi | 0.045 | 0.104 | 0.269 | boundary | MID|LCD1; | 0.0 |
| 72 | fp_D | Q04377 | 685 | AFEDLPEYIEEELKMQL ................. | HHHCCHHHHHHHHHHHH | c1c-5 | uniq | 0.131 | 0.067 | 0.268 | boundary | MID|LCD1; | 0.0 |
| 73 | fp_D | Q04377 | 697 | LKMQLNKRTGRIMQVKY ................. | HHHHHHHHCCCCCCCCC | c1c-AT-4 | uniq | 0.158 | 0.075 | 0.29 | boundary | MID|LCD1; | 0.0 |
| 74 | fp_D | Q04377 | 731 | DLTTLEEADSLYISM ............... | CCCCHHHHHHHHHHC | c1b-AT-5 | multi-selected | 0.166 | 0.097 | 0.181 | boundary | boundary|LCD1; | 0.0 |
| 75 | fp_D | Q04377 | 734 | TLEEADSLYISMGLXX .............. | CHHHHHHHHHHCCC | c1d-AT-5-Ct | multi-selected | 0.175 | 0.127 | 0.172 | boundary | boundary|LCD1; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment