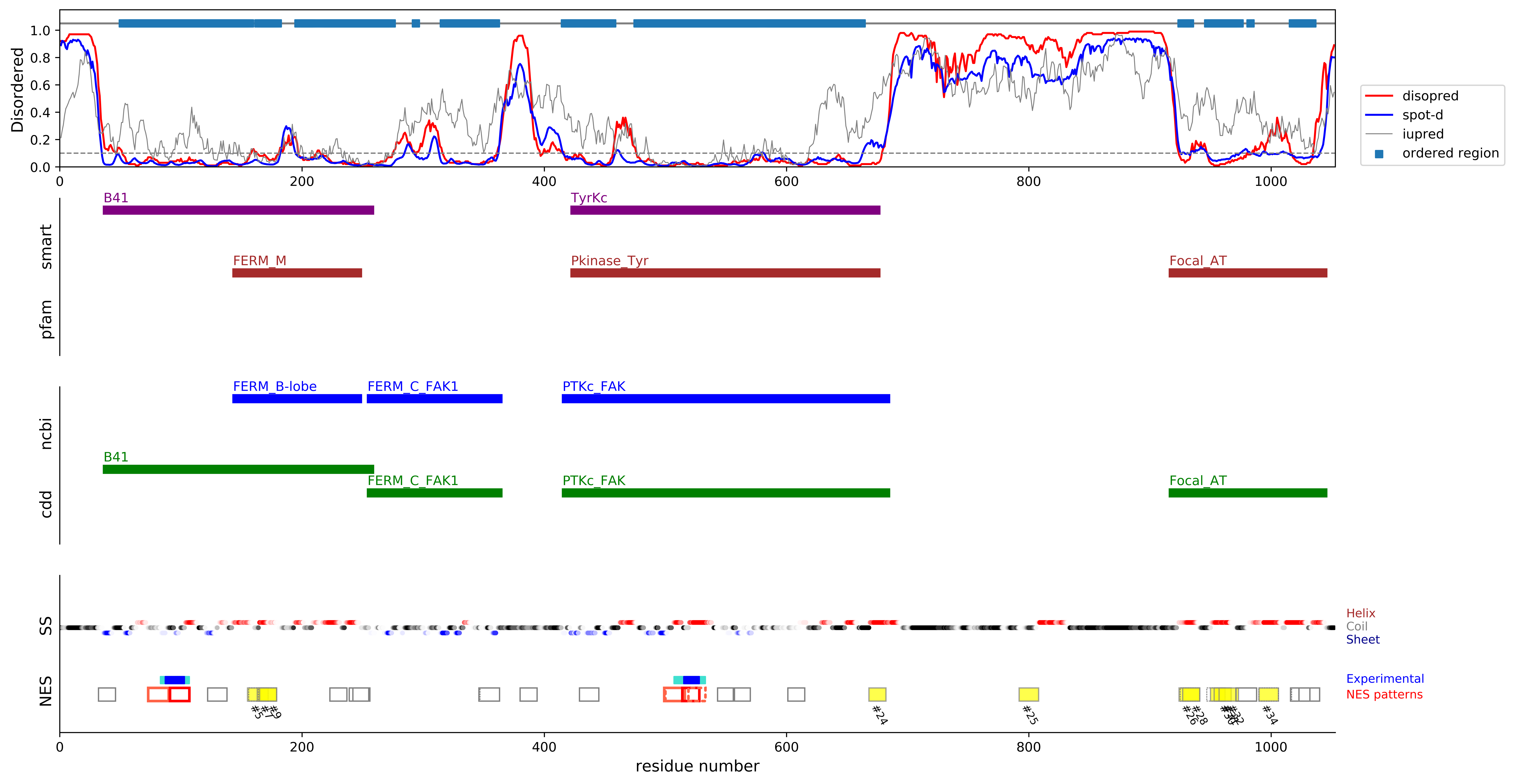

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q05397 | 32 | AMERVLKVFHYFES | CCCCEEEEEEEECC | c3-4 | uniq | 0.226 | 0.077 | 0.252 | boundary | boundary|B41; | 0.857 |
| 2 | cand_O | Q05397 | 73 | DSHKVKHVACYGFRLSH ++++ | CCCCCCCCCCCEEEECC | c1c-AT-4 | uniq | 0.045 | 0.024 | 0.11 | ORD | MID|B41; | 0.125 |
| 3 | cand_beta_O | Q05397 | 91 | RSEEVHWLHVDMGVSS ++++*++*+*++++ | CCCCEEEECCCCCHHH | c1d-4 | uniq | 0.043 | 0.024 | 0.172 | ORD | MID|B41; | 0.571 |
| 4 | fp_O | Q05397 | 122 | YELRIRYLPKGFLNQF | EEEEEEECCHHHHHHH | c1aR-4 | uniq | 0.017 | 0.02 | 0.113 | ORD | MID|B41; | 0.429 |
| 5 | fp_D | Q05397 | 155 | YMLEIADQVDQEIALKL | HHHHHCCCCCHHHHHHH | c1c-5 | multi-selected | 0.089 | 0.034 | 0.074 | boundary | MID|B41; | 0.0 |
| 6 | fp_D | Q05397 | 156 | MLEIADQVDQEIALKL | HHHHCCCCCHHHHHHH | c1d-AT-5 | multi | 0.092 | 0.035 | 0.074 | boundary | MID|B41; | 0.0 |
| 7 | fp_D | Q05397 | 163 | VDQEIALKLGCLEIRR | CCHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.071 | 0.025 | 0.047 | boundary | MID|B41; | 0.0 |
| 8 | fp_D | Q05397 | 164 | DQEIALKLGCLEIRR | CHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.069 | 0.023 | 0.046 | boundary | MID|B41; | 0.0 |
| 9 | fp_D | Q05397 | 165 | QEIALKLGCLEIRR | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.067 | 0.022 | 0.047 | boundary | MID|B41; | 0.0 |
| 10 | fp_O | Q05397 | 223 | LIQQTFRQFANLNR | HHHHHHHHHCCCCH | c3-AT | uniq | 0.031 | 0.033 | 0.128 | ORD | MID|B41; boundary|FERM_C_FAK1; | 0.0 |
| 11 | fp_O | Q05397 | 239 | SILKFFEILSPVYRFD | HHHHHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.019 | 0.015 | 0.037 | ORD | boundary|B41; boundary|FERM_C_FAK1; | 0.0 |
| 12 | fp_O | Q05397 | 242 | KFFEILSPVYRFDK | HHHHHHCCCCCCCC | c3-4 | multi-selected | 0.016 | 0.015 | 0.033 | ORD | boundary|B41; boundary|FERM_C_FAK1; | 0.0 |
| 13 | fp_D | Q05397 | 346 | GYCRLVNGTSQSFIIRP | CCCCCCCCCCCEEEECC | c1c-AT-4 | multi-selected | 0.049 | 0.044 | 0.298 | boundary | boundary|FERM_C_FAK1; | 0.125 |
| 14 | fp_D | Q05397 | 347 | YCRLVNGTSQSFIIRP | CCCCCCCCCCEEEECC | c1d-AT-4 | multi | 0.051 | 0.045 | 0.301 | boundary | boundary|FERM_C_FAK1; | 0.143 |
| 15 | fp_D | Q05397 | 380 | EKQGMRTHAVSVSE | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.679 | 0.532 | 0.548 | DISO | boundary|FERM_C_FAK1; | 0.0 |
| 16 | fp_beta_D | Q05397 | 429 | GEGQFGDVHQGIYMSP | CCCCEEEEEEEEEECC | c1d-4 | uniq | 0.043 | 0.031 | 0.268 | boundary | boundary|PTKc_FAK; | 1.0 |
| 17 | cand_O | Q05397 | 499 | MELCTLGELRSFLQVRK ++++++ | EEECCCCHHHHHHHHCC | c1c-AT-4 | multi-selected | 0.016 | 0.025 | 0.026 | ORD | MID|PTKc_FAK; | 0.0 |
| 18 | cand_O | Q05397 | 500 | ELCTLGELRSFLQVRK ++++++ | EECCCCHHHHHHHHCC | c1d-5 | multi | 0.016 | 0.026 | 0.024 | ORD | MID|PTKc_FAK; | 0.0 |
| 19 | cand_O | Q05397 | 514 | RKYSLDLASLILYA ++++*+*++*+*++ | CCCCCCHHHHHHHH | c2-4 | multi-selected | 0.023 | 0.02 | 0.011 | ORD | MID|PTKc_FAK; | 0.0 |
| 20 | cand_O | Q05397 | 519 | DLASLILYAYQLST +*++*+*+++++ | CHHHHHHHHHHHHH | c3-AT | multi | 0.021 | 0.009 | 0.01 | ORD | MID|PTKc_FAK; | 0.0 |
| 21 | fp_O | Q05397 | 543 | VHRDIAARNVLVSS | CCCCCCCCEEECCC | c2-AT-4 | uniq | 0.016 | 0.021 | 0.085 | ORD | MID|PTKc_FAK; | 0.286 |
| 22 | fp_beta_O | Q05397 | 556 | SNDCVKLGDFGLSR | CCCEEEEECCCCCE | c2-4 | uniq | 0.039 | 0.018 | 0.07 | ORD | MID|PTKc_FAK; | 0.714 |
| 23 | fp_O | Q05397 | 601 | SASDVWMFGVCMWE | CCCCCCCCCCHHHH | c2-4 | uniq | 0.021 | 0.031 | 0.028 | ORD | MID|PTKc_FAK; | 0.0 |
| 24 | fp_D | Q05397 | 668 | RFTELKAQLSTILE | CHHHHHHHHHHHHH | c3-4 | uniq | 0.051 | 0.165 | 0.499 | boundary | boundary|PTKc_FAK; | 0.0 |
| 25 | fp_D | Q05397 | 792 | TVLDLRGIGQVLPTHL | CCCCCCCCCCCCCCCH | c1aR-4 | uniq | 0.896 | 0.742 | 0.54 | DISO | 0.0 | |
| 26 | fp_D | Q05397 | 924 | VYENVTGLVKAVIEMSS | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.094 | 0.108 | 0.389 | boundary | boundary|Focal_AT; | 0.0 |
| 27 | fp_D | Q05397 | 925 | YENVTGLVKAVIEMSS | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.094 | 0.106 | 0.383 | boundary | boundary|Focal_AT; | 0.0 |
| 28 | fp_D | Q05397 | 927 | NVTGLVKAVIEMSS | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.099 | 0.107 | 0.382 | boundary | boundary|Focal_AT; | 0.0 |
| 29 | fp_D | Q05397 | 947 | PEEYVPMVKEVGLAL | CCCCHHHHHHHHHHH | c1b-4 | multi | 0.027 | 0.056 | 0.399 | boundary | MID|Focal_AT; | 0.0 |
| 30 | fp_D | Q05397 | 950 | YVPMVKEVGLAL | CHHHHHHHHHHH | c2-rev | multi-selected | 0.017 | 0.049 | 0.36 | boundary | MID|Focal_AT; | 0.0 |
| 31 | fp_D | Q05397 | 953 | MVKEVGLALRTLLA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.02 | 0.052 | 0.311 | boundary | MID|Focal_AT; | 0.0 |
| 32 | fp_D | Q05397 | 957 | VGLALRTLLATVDETI | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.033 | 0.065 | 0.301 | boundary | MID|Focal_AT; | 0.0 |
| 33 | fp_D | Q05397 | 971 | TIPLLPASTHREIEMAQ | CCCCCCCCCCCHHHHHH | c1c-AT-5 | uniq | 0.083 | 0.104 | 0.448 | boundary | MID|Focal_AT; | 0.0 |
| 34 | fp_D | Q05397 | 990 | LNSDLGELINKMKLAQ | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.207 | 0.095 | 0.275 | boundary | MID|Focal_AT; | 0.0 |
| 35 | fp_D | Q05397 | 990 | LNSDLGELINKMKLAQ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.207 | 0.095 | 0.275 | boundary | MID|Focal_AT; | 0.0 |
| 36 | fp_D | Q05397 | 1016 | YKKQMLTAAHALAVDA | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi-selected | 0.043 | 0.071 | 0.225 | boundary | boundary|Focal_AT; | 0.0 |
| 37 | fp_D | Q05397 | 1016 | YKKQMLTAAHALAVDA | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.043 | 0.071 | 0.225 | boundary | boundary|Focal_AT; | 0.0 |
| 38 | fp_D | Q05397 | 1017 | KKQMLTAAHALAVDA | HHHHHHHHHHHHHCC | c1b-AT-4 | multi | 0.039 | 0.07 | 0.221 | boundary | boundary|Focal_AT; | 0.0 |
| 39 | fp_D | Q05397 | 1023 | AAHALAVDAKNLLDVID | HHHHHHHCCCCHHHHHH | c1c-AT-4 | uniq | 0.1 | 0.07 | 0.18 | boundary | boundary|Focal_AT; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment