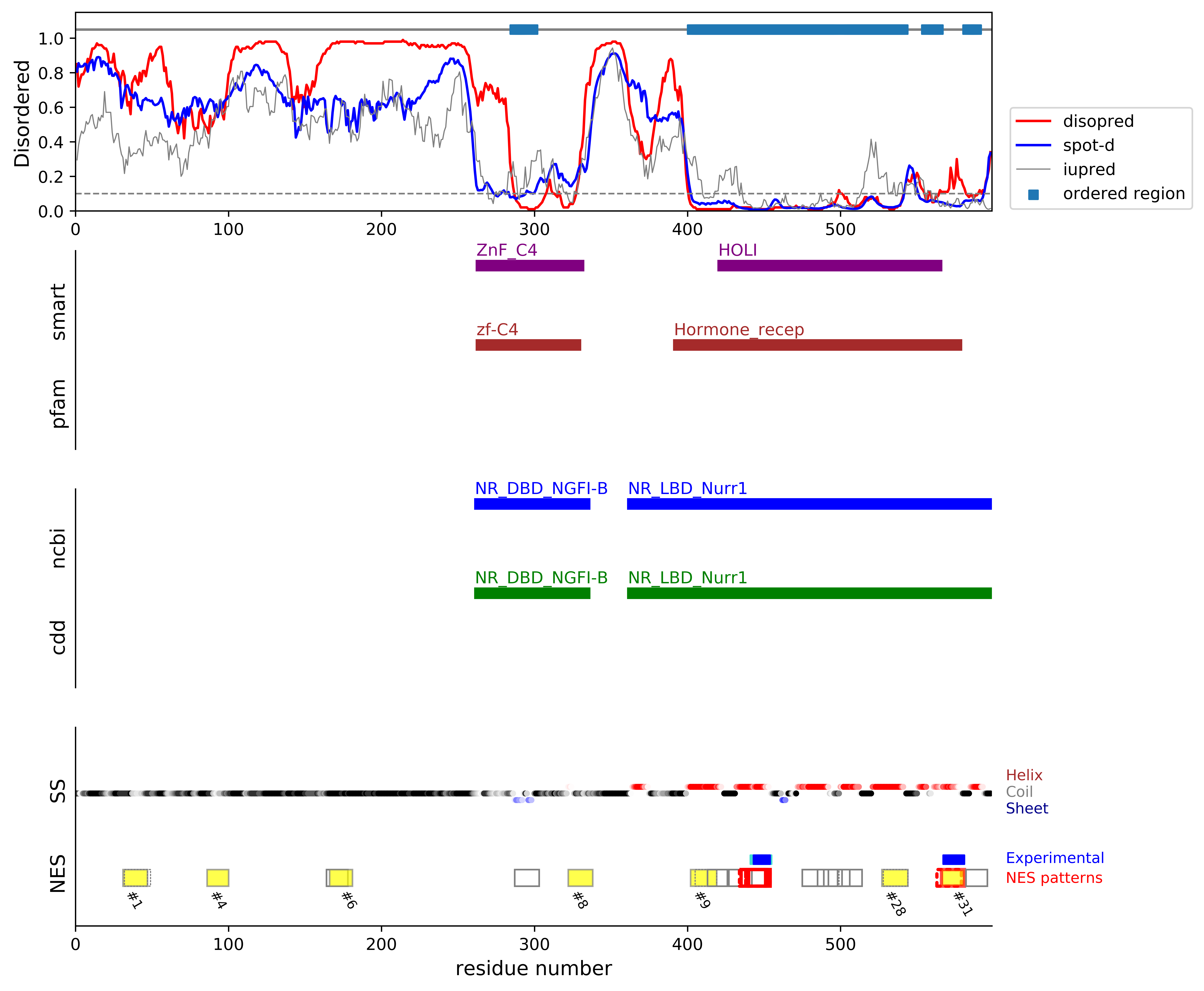

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q06219 | 31 | SSDFLTPEFVKFSMDL | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.736 | 0.66 | 0.334 | DISO | 0.0 | |
| 2 | fp_D | Q06219 | 32 | SDFLTPEFVKFSMDL | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.731 | 0.662 | 0.336 | DISO | 0.0 | |
| 3 | fp_D | Q06219 | 32 | SDFLTPEFVKFSMDLTN | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi | 0.746 | 0.659 | 0.344 | DISO | 0.0 | |
| 4 | fp_D | Q06219 | 86 | QQSSIKVEDIQMHN | CCCCCCCCCCCCCC | c2-4 | uniq | 0.557 | 0.616 | 0.541 | DISO | 0.0 | |
| 5 | fp_D | Q06219 | 164 | EQRKTPVSRLSLFS | CCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.966 | 0.576 | 0.519 | DISO | 0.0 | |
| 6 | fp_D | Q06219 | 166 | RKTPVSRLSLFSFKQ | CCCCCCCCCCCCCCC | c1b-4 | multi-selected | 0.973 | 0.578 | 0.511 | DISO | 0.0 | |
| 7 | fp_beta_D | Q06219 | 287 | FKRTVQKNAKYVCLAN | EEEEEECCCEEECCCC | c1a-AT-4 | uniq | 0.027 | 0.098 | 0.2 | boundary | MID|NR_DBD_NGFI-B; | 0.571 |
| 8 | fp_D | Q06219 | 322 | KCLAVGMVKEVVRTDS | HHHCCCCCCCCCCCCC | c1aR-4 | uniq | 0.341 | 0.243 | 0.234 | DISO | boundary|NR_DBD_NGFI-B; | 0.0 |
| 9 | fp_D | Q06219 | 402 | HIQQFYDLLTGSMEIIR | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.014 | 0.049 | 0.159 | boundary | MID|NR_LBD_Nurr1; | 0.0 |
| 10 | fp_D | Q06219 | 405 | QFYDLLTGSMEIIR | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.011 | 0.043 | 0.133 | boundary | MID|NR_LBD_Nurr1; | 0.0 |
| 11 | fp_D | Q06219 | 413 | SMEIIRGWAEKIPG | HHHHHHHHHHCCCC | c3-AT | uniq | 0.01 | 0.048 | 0.141 | boundary | MID|NR_LBD_Nurr1; | 0.0 |
| 12 | fp_O | Q06219 | 426 | GFADLPKADQDLLFES | CCCCCCHHHHHHHHHH | c1d-AT-5 | uniq | 0.019 | 0.027 | 0.139 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 13 | cand_O | Q06219 | 434 | DQDLLFESAFLELFVLR ++*++*+ | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.015 | 0.01 | 0.046 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 14 | fp_O | Q06219 | 435 | QDLLFESAFLELFVLRL ++*++*+* | HHHHHHHHHHHHHHHHH | c1cR-4 | multi | 0.014 | 0.01 | 0.044 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 15 | cand_O | Q06219 | 435 | QDLLFESAFLELFVLRL ++*++*+* | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.014 | 0.01 | 0.044 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 16 | cand_O | Q06219 | 435 | QDLLFESAFLELFVLR ++*++*+ | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.014 | 0.01 | 0.043 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 17 | cand_O | Q06219 | 435 | QDLLFESAFLELFVLR ++*++*+ | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.014 | 0.01 | 0.043 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 18 | cand_O | Q06219 | 438 | LFESAFLELFVLRLAY ++*++*+*+ | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.013 | 0.009 | 0.032 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 19 | cand_O | Q06219 | 438 | LFESAFLELFVLRL ++*++*+* | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.013 | 0.008 | 0.033 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 20 | cand_O | Q06219 | 439 | FESAFLELFVLRLAY ++*++*+*+ | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.012 | 0.009 | 0.031 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 21 | cand_O | Q06219 | 440 | ESAFLELFVLRLAY ++*++*+*+ | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.011 | 0.008 | 0.03 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 22 | fp_O | Q06219 | 475 | CVRGFGEWIDSIVEFSS | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.014 | 0.016 | 0.044 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 23 | fp_O | Q06219 | 475 | CVRGFGEWIDSIVE | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.013 | 0.016 | 0.04 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 24 | fp_O | Q06219 | 485 | SIVEFSSNLQNMNIDI | HHHHHHHHCCCCCCCH | c1a-5 | uniq | 0.041 | 0.024 | 0.051 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 25 | fp_O | Q06219 | 492 | NLQNMNIDISAFSC | HCCCCCCCHHHHHH | c3-4 | uniq | 0.067 | 0.026 | 0.034 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 26 | fp_O | Q06219 | 498 | IDISAFSCIAALAMVT | CCHHHHHHHHHHHHHC | c1a-AT-4 | multi-selected | 0.059 | 0.018 | 0.038 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 27 | fp_O | Q06219 | 499 | DISAFSCIAALAMVT | CHHHHHHHHHHHHHC | c1b-5 | multi | 0.057 | 0.017 | 0.038 | ORD | MID|NR_LBD_Nurr1; | 0.0 |
| 28 | fp_D | Q06219 | 527 | LQNKIVNCLKDHVTFNN | HHHHHHHHHHHHHHHCC | c1c-4 | multi-selected | 0.041 | 0.044 | 0.144 | boundary | MID|NR_LBD_Nurr1; | 0.0 |
| 29 | fp_D | Q06219 | 528 | QNKIVNCLKDHVTFNN | HHHHHHHHHHHHHHCC | c1d-4 | multi | 0.042 | 0.045 | 0.14 | boundary | MID|NR_LBD_Nurr1; | 0.0 |
| 30 | cand_D | Q06219 | 563 | RTLCTQGLQRIFYLKL *++*++*+* | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.15 | 0.039 | 0.044 | boundary | boundary|NR_LBD_Nurr1; | 0.0 |
| 31 | cand_D | Q06219 | 566 | CTQGLQRIFYLKLED *++*++*+* | HHHHHHHHHHHHHCC | c1b-4 | multi-selected | 0.163 | 0.036 | 0.041 | boundary | boundary|NR_LBD_Nurr1; | 0.0 |
| 32 | fp_D | Q06219 | 582 | VPPPAIIDKLFLDT | CCCHHHHHHHHHCC | c2-AT-4 | uniq | 0.101 | 0.072 | 0.044 | __ | boundary|NR_LBD_Nurr1; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment