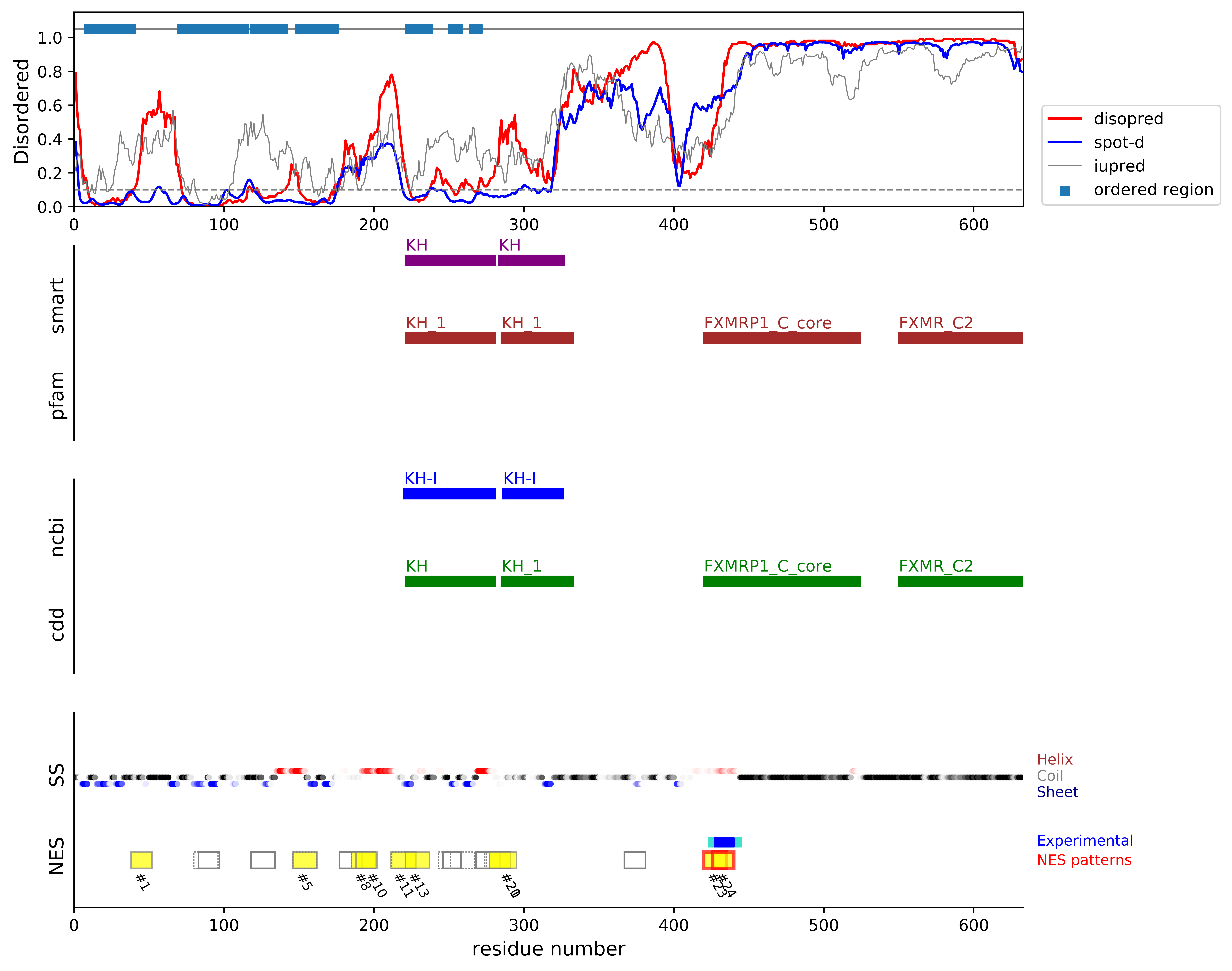

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q06787 | 38 | PDRQIPFHDVRFPP | CCCCCCCCCEECCC | c2-4 | uniq | 0.324 | 0.046 | 0.341 | boundary | 0.143 | |
| 2 | fp_beta_D | Q06787 | 80 | WLAKVRMIKGEFYVIE | EEEEEEEEECCEEEEE | c1d-5 | multi | 0.01 | 0.01 | 0.047 | boundary | 0.714 | |
| 3 | fp_beta_D | Q06787 | 83 | KVRMIKGEFYVIEY | EEEEEECCEEEEEE | c3-4 | multi-selected | 0.01 | 0.011 | 0.034 | boundary | 0.714 | |
| 4 | fp_D | Q06787 | 118 | NKPATKDTFHKIKLDV | CCCCCCCCEEEEEECC | c1a-AT-4 | uniq | 0.073 | 0.059 | 0.413 | boundary | 0.429 | |
| 5 | fp_D | Q06787 | 146 | AHKDFKKAVGAFSVTY | HHHHHHHHHCCEEEEE | c1a-4 | multi-selected | 0.089 | 0.021 | 0.207 | boundary | 0.0 | |
| 6 | fp_D | Q06787 | 146 | AHKDFKKAVGAFSVTY | HHHHHHHHHCCEEEEE | c1d-AT-4 | multi | 0.089 | 0.021 | 0.207 | boundary | 0.0 | |
| 7 | fp_D | Q06787 | 177 | TSKRAHMLIDMHFRS | CHHHHCCCCCCCCCH | c1b-AT-4 | uniq | 0.278 | 0.209 | 0.172 | boundary | 0.0 | |
| 8 | fp_D | Q06787 | 185 | IDMHFRSLRTKLSLIM | CCCCCCHHHHHHHHHH | c1aR-5 | multi-selected | 0.32 | 0.249 | 0.207 | boundary | boundary|KH; | 0.0 |
| 9 | fp_D | Q06787 | 185 | IDMHFRSLRTKLSLIM | CCCCCCHHHHHHHHHH | c1d-4 | multi | 0.32 | 0.249 | 0.207 | boundary | boundary|KH; | 0.0 |
| 10 | fp_D | Q06787 | 188 | HFRSLRTKLSLIMR | CCCHHHHHHHHHHH | c3-4 | multi-selected | 0.323 | 0.261 | 0.211 | DISO | boundary|KH; | 0.0 |
| 11 | fp_D | Q06787 | 211 | SSRQLASRFHEQFIVRE | HHHHHCCCCEEEEEECC | c1c-4 | multi-selected | 0.376 | 0.164 | 0.267 | boundary | boundary|KH; | 0.375 |
| 12 | fp_D | Q06787 | 212 | SRQLASRFHEQFIVRE | HHHHCCCCEEEEEECC | c1d-AT-4 | multi | 0.351 | 0.151 | 0.249 | boundary | boundary|KH; | 0.429 |
| 13 | fp_D | Q06787 | 221 | EQFIVREDLMGLAIGT | EEEEECCCHHHHHCCC | c1a-4 | uniq | 0.073 | 0.057 | 0.205 | boundary | boundary|KH; | 0.143 |
| 14 | fp_beta_D | Q06787 | 243 | QARKVPGVTAIDLDE | HHHCCCCEEEEEECC | c1b-4 | multi | 0.13 | 0.059 | 0.381 | boundary | MID|KH; | 0.571 |
| 15 | fp_beta_D | Q06787 | 246 | KVPGVTAIDLDE | CCCCEEEEEECC | c2-rev | multi-selected | 0.113 | 0.049 | 0.375 | boundary | MID|KH; | 0.833 |
| 16 | fp_D | Q06787 | 251 | TAIDLDEDTCTFHIYG | EEEEECCCCCEEEEEE | c1a-AT-4 | multi | 0.112 | 0.031 | 0.355 | boundary | boundary|KH; | 0.286 |
| 17 | fp_D | Q06787 | 268 | DQDAVKKARSFLEFAE | CHHHHHHHHHHHHCCE | c1d-AT-4 | uniq | 0.159 | 0.069 | 0.286 | boundary | boundary|KH; | 0.0 |
| 18 | fp_D | Q06787 | 274 | KARSFLEFAEDVIQVPR | HHHHHHHCCEEECCCCC | c1c-AT-4 | multi | 0.284 | 0.063 | 0.258 | DISO | boundary|KH; | 0.375 |
| 19 | fp_D | Q06787 | 275 | ARSFLEFAEDVIQVPR | HHHHHHCCEEECCCCC | c1d-AT-4 | multi | 0.294 | 0.063 | 0.255 | DISO | boundary|KH; | 0.429 |
| 20 | fp_D | Q06787 | 277 | SFLEFAEDVIQVPR | HHHHCCEEECCCCC | c3-4 | multi-selected | 0.319 | 0.061 | 0.244 | DISO | boundary|KH; | 0.429 |
| 21 | fp_D | Q06787 | 277 | SFLEFAEDVIQVPRNLVG | HHHHCCEEECCCCCCCCC | c4-5 | multi-selected | 0.357 | 0.065 | 0.26 | DISO | boundary|KH; | 0.375 |
| 22 | fp_D | Q06787 | 367 | QPNSTKVQRVLVAS | CCCCCCCEEEEEEC | c2-AT-4 | uniq | 0.869 | 0.613 | 0.488 | DISO | 0.429 | |
| 23 | cand_D | Q06787 | 420 | HLNYLKEVDQLRLER ++++*+*++ | HHHHHHHHHHHHHHH | c1b-5 | uniq | 0.505 | 0.62 | 0.384 | DISO | boundary|FXMRP1_C_core; | 0.0 |
| 24 | cand_D | Q06787 | 426 | EVDQLRLERLQIDE ++++*+*++*+*++ | HHHHHHHHHHHHHH | c2-5 | uniq | 0.714 | 0.659 | 0.456 | DISO | boundary|FXMRP1_C_core; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment