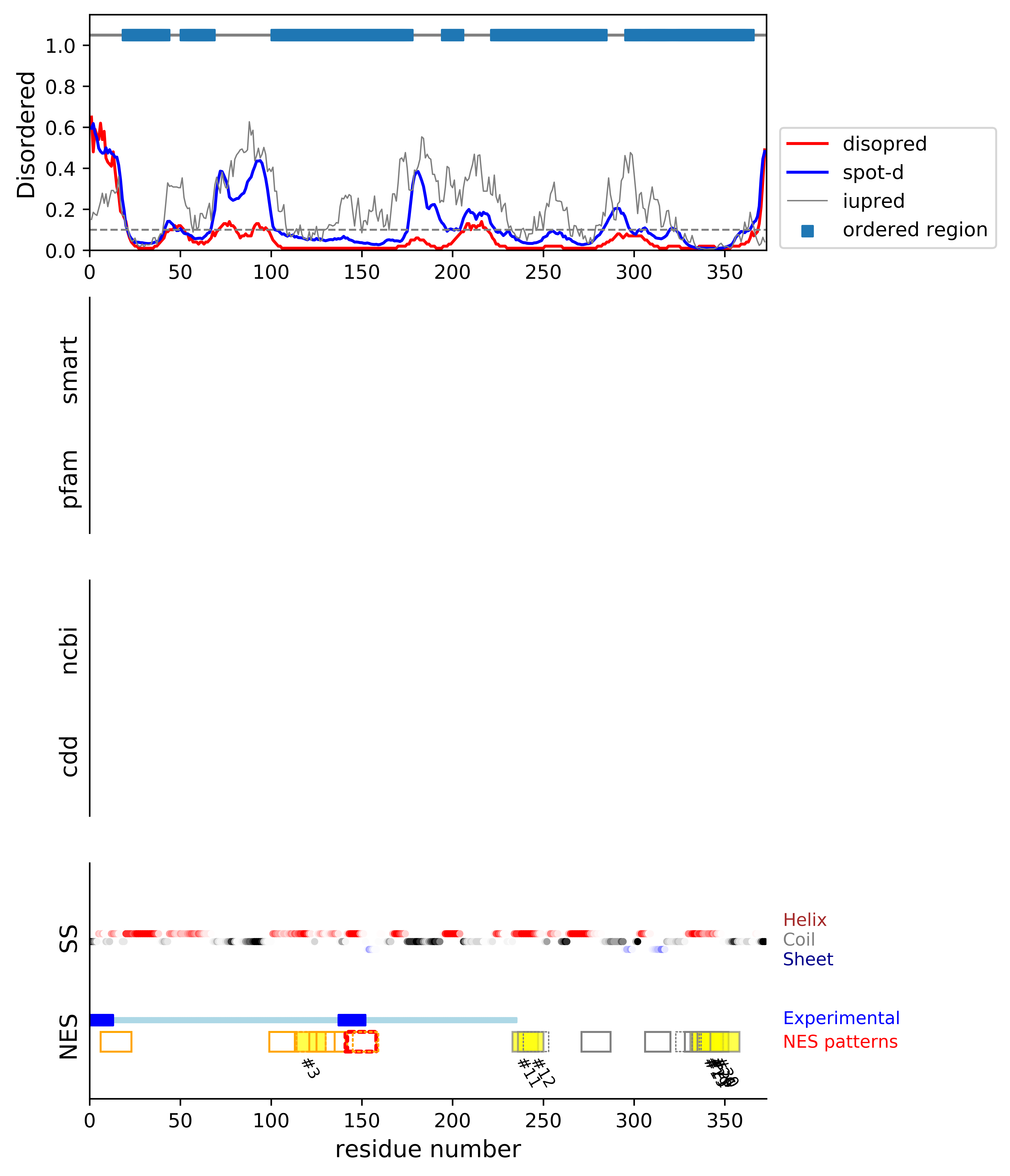

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q08358 | 6 | EELGLSKAARSQVDLNQ ..*.*............ | HHHCCCHHHHHHCCHHH | c1c-AT-4 | uniq | 0.336 | 0.367 | 0.227 | boundary | 0.0 | |
| 2 | fp_D | Q08358 | 99 | ADWKAAVSAIELEY .............. | CCHHHHHHHHHHHH | c2-AT-4 | uniq | 0.028 | 0.102 | 0.213 | boundary | 0.0 | |
| 3 | fp_D | Q08358 | 114 | VKRRFYRALEGLDLYL ................ | HHHHHHHHHCCHHHHH | c1a-4 | multi-selected | 0.01 | 0.056 | 0.076 | boundary | 0.0 | |
| 4 | fp_D | Q08358 | 114 | VKRRFYRALEGLDLYL ................ | HHHHHHHHHCCHHHHH | c1d-AT-4 | multi | 0.01 | 0.056 | 0.076 | boundary | 0.0 | |
| 5 | fp_O | Q08358 | 121 | ALEGLDLYLKNITK .............. | HHCCHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.052 | 0.089 | ORD | 0.0 | |
| 6 | fp_O | Q08358 | 125 | LDLYLKNITKTFVNNI ...............* | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.01 | 0.054 | 0.129 | ORD | 0.0 | |
| 7 | cand_O | Q08358 | 141 | DSIQTVQQMLDGVRIIG ..*..*..*........ | CHHHHHHHHHHCEEEEE | c1c-AT-4 | multi | 0.01 | 0.04 | 0.195 | ORD | 0.0 | |
| 8 | fp_O | Q08358 | 142 | SIQTVQQMLDGVRIIG .*..*..*........ | HHHHHHHHHHCEEEEE | c1a-5 | multi-selected | 0.01 | 0.039 | 0.19 | ORD | 0.0 | |
| 9 | cand_O | Q08358 | 142 | SIQTVQQMLDGVRIIG .*..*..*........ | HHHHHHHHHHCEEEEE | c1d-5 | multi | 0.01 | 0.039 | 0.19 | ORD | 0.0 | |
| 10 | fp_O | Q08358 | 145 | TVQQMLDGVRIIGR .*..*......... | HHHHHHHCEEEEEE | c3-4 | multi | 0.01 | 0.035 | 0.174 | ORD | 0.286 | |
| 11 | fp_D | Q08358 | 233 | NFQALKNIINAFAR .. | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.043 | 0.086 | boundary | 0.0 | |
| 12 | fp_D | Q08358 | 236 | ALKNIINAFARIGD | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.039 | 0.102 | boundary | 0.0 | |
| 13 | fp_O | Q08358 | 239 | NIINAFARIGDMLG | HHHHHHHHHHHCCC | c3-AT | multi | 0.011 | 0.043 | 0.123 | ORD | 0.0 | |
| 14 | fp_D | Q08358 | 271 | LLEYIQHSALSVGLKN | HHHHHHHHHHHHCCCC | c1a-AT-5 | uniq | 0.018 | 0.067 | 0.12 | boundary | 0.0 | |
| 15 | fp_beta_D | Q08358 | 306 | AAQRVYLSTVRVND | HHHHEEEEEEEECC | c2-4 | uniq | 0.024 | 0.074 | 0.202 | boundary | 0.857 | |
| 16 | fp_D | Q08358 | 323 | TRWETEDVFFTFMLKS | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi | 0.014 | 0.037 | 0.049 | boundary | 0.0 | |
| 17 | fp_D | Q08358 | 323 | TRWETEDVFFTFMLKS | CCCCCCHHHHHHHHHH | c1d-AT-4 | multi | 0.014 | 0.037 | 0.049 | boundary | 0.0 | |
| 18 | fp_O | Q08358 | 328 | EDVFFTFMLKSMAAKI | CHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.015 | 0.016 | 0.026 | ORD | 0.0 | |
| 19 | fp_D | Q08358 | 331 | FFTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.015 | 0.011 | 0.018 | boundary | 0.0 | |
| 20 | fp_D | Q08358 | 332 | FTFMLKSMAAKIFIVLG | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.015 | 0.011 | 0.017 | boundary | 0.0 | |
| 21 | fp_D | Q08358 | 332 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.016 | 0.011 | 0.016 | boundary | 0.0 | |
| 22 | fp_D | Q08358 | 332 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.016 | 0.011 | 0.016 | boundary | 0.0 | |
| 23 | fp_D | Q08358 | 332 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.016 | 0.011 | 0.016 | boundary | 0.0 | |
| 24 | fp_D | Q08358 | 335 | MLKSMAAKIFIVLGIYD | HHHHHHHHHHHHHHCCC | c1c-5 | multi-selected | 0.015 | 0.011 | 0.013 | boundary | 0.0 | |
| 25 | fp_D | Q08358 | 335 | MLKSMAAKIFIVLG | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.016 | 0.01 | 0.012 | boundary | 0.0 | |
| 26 | fp_D | Q08358 | 335 | MLKSMAAKIFIVLG | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.016 | 0.01 | 0.012 | boundary | 0.0 | |
| 27 | fp_D | Q08358 | 336 | LKSMAAKIFIVLGIYD | HHHHHHHHHHHHHCCC | c1a-AT-4 | multi | 0.016 | 0.011 | 0.013 | boundary | 0.0 | |
| 28 | fp_D | Q08358 | 336 | LKSMAAKIFIVLGIYD | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.016 | 0.011 | 0.013 | boundary | 0.0 | |
| 29 | fp_D | Q08358 | 337 | KSMAAKIFIVLGIYD | HHHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.015 | 0.011 | 0.013 | boundary | 0.0 | |
| 30 | fp_D | Q08358 | 342 | KIFIVLGIYDMFERPE | HHHHHHHCCCCCCCCC | c1aR-4 | multi-selected | 0.014 | 0.023 | 0.024 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment