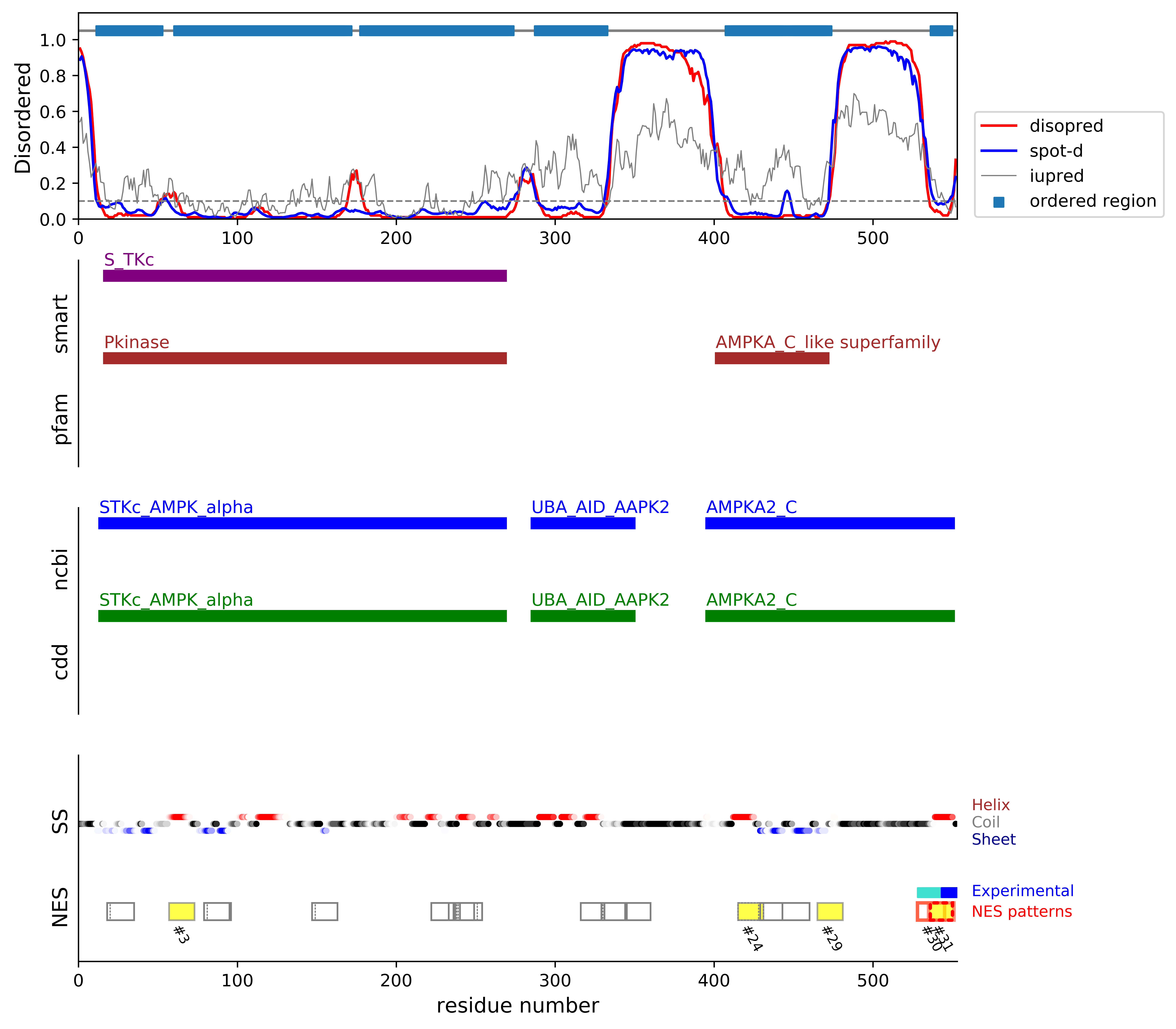

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q09137 | 18 | LGDTLGVGTFGKVKIGE | EEEEEECCCCEEEEEEE | c1c-AT-4 | multi-selected | 0.021 | 0.066 | 0.134 | boundary | boundary|STKc_AMPK_alpha; | 0.5 |
| 2 | fp_D | Q09137 | 20 | DTLGVGTFGKVKIGE | EEEECCCCEEEEEEE | c1b-4 | multi | 0.021 | 0.066 | 0.138 | boundary | boundary|STKc_AMPK_alpha; | 0.429 |
| 3 | fp_D | Q09137 | 57 | VVGKIKREIQNLKLFR | HHHHHHHHHHHHHHCC | c1a-5 | uniq | 0.069 | 0.038 | 0.129 | boundary | MID|STKc_AMPK_alpha; | 0.0 |
| 4 | fp_beta_O | Q09137 | 79 | LYQVISTPTDFFMVME | EEEEEECCCEEEEEEE | c1a-AT-4 | multi-selected | 0.009 | 0.009 | 0.054 | ORD | MID|STKc_AMPK_alpha; | 0.571 |
| 5 | fp_beta_O | Q09137 | 79 | LYQVISTPTDFFMVMEY | EEEEEECCCEEEEEEEC | c1c-AT-4 | multi-selected | 0.008 | 0.009 | 0.053 | ORD | MID|STKc_AMPK_alpha; | 0.625 |
| 6 | fp_beta_O | Q09137 | 81 | QVISTPTDFFMVME | EEEECCCEEEEEEE | c3-AT | multi | 0.009 | 0.009 | 0.053 | ORD | MID|STKc_AMPK_alpha; | 0.571 |
| 7 | fp_beta_D | Q09137 | 147 | LDAQMNAKIADFGLSN | CCCCCCEEEECCCCCC | c1a-4 | multi-selected | 0.01 | 0.02 | 0.113 | boundary | MID|STKc_AMPK_alpha; | 0.571 |
| 8 | fp_beta_D | Q09137 | 149 | AQMNAKIADFGLSN | CCCCEEEECCCCCC | c2-AT-4 | multi | 0.01 | 0.02 | 0.105 | boundary | MID|STKc_AMPK_alpha; | 0.571 |
| 9 | fp_O | Q09137 | 222 | LFKKIRGGVFYIPE | HHHHHHCCEECCCC | c3-4 | multi-selected | 0.01 | 0.032 | 0.054 | ORD | MID|STKc_AMPK_alpha; | 0.286 |
| 10 | fp_O | Q09137 | 222 | LFKKIRGGVFYIPEYLNR | HHHHHHCCEECCCCCCCH | c4-5 | multi-selected | 0.01 | 0.033 | 0.048 | ORD | MID|STKc_AMPK_alpha; | 0.25 |
| 11 | fp_O | Q09137 | 233 | IPEYLNRSIATLLMHM | CCCCCCHHHHHHHHHH | c1a-4 | multi-selected | 0.01 | 0.034 | 0.055 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 12 | fp_O | Q09137 | 237 | LNRSIATLLMHMLQ | CCHHHHHHHHHHCC | c2-AT-4 | multi | 0.01 | 0.034 | 0.066 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 13 | fp_O | Q09137 | 237 | LNRSIATLLMHMLQVDP | CCHHHHHHHHHHCCCCC | c1c-4 | multi-selected | 0.01 | 0.039 | 0.072 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 14 | fp_O | Q09137 | 238 | NRSIATLLMHMLQVDP | CHHHHHHHHHHCCCCC | c1a-AT-4 | multi | 0.01 | 0.039 | 0.075 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 15 | fp_O | Q09137 | 238 | NRSIATLLMHMLQVDP | CHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.01 | 0.039 | 0.075 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 16 | fp_O | Q09137 | 239 | RSIATLLMHMLQVDP | HHHHHHHHHHCCCCC | c1b-AT-4 | multi | 0.01 | 0.039 | 0.077 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 17 | fp_O | Q09137 | 240 | SIATLLMHMLQVDP | HHHHHHHHHCCCCC | c2-5 | multi-selected | 0.01 | 0.039 | 0.082 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 18 | fp_O | Q09137 | 240 | SIATLLMHMLQVDP | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.01 | 0.039 | 0.082 | ORD | boundary|STKc_AMPK_alpha; | 0.0 |
| 19 | fp_D | Q09137 | 316 | PQDQLAVAYHLIIDN | CCCHHHHHHHHHHHC | c1b-AT-4 | uniq | 0.013 | 0.052 | 0.205 | boundary | boundary|UBA_AID_AAPK2; | 0.0 |
| 20 | fp_D | Q09137 | 329 | DNRRIMNQASEFYLAS | HCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.472 | 0.498 | 0.231 | boundary | boundary|UBA_AID_AAPK2; | 0.0 |
| 21 | fp_D | Q09137 | 330 | NRRIMNQASEFYLAS | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.503 | 0.527 | 0.234 | boundary | boundary|UBA_AID_AAPK2; | 0.0 |
| 22 | fp_D | Q09137 | 344 | SSPPTGSFMDDMAMHI | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.964 | 0.931 | 0.408 | boundary | boundary|UBA_AID_AAPK2; | 0.0 |
| 23 | fp_D | Q09137 | 344 | SSPPTGSFMDDMAMHI | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.964 | 0.931 | 0.408 | boundary | boundary|UBA_AID_AAPK2; | 0.0 |
| 24 | fp_D | Q09137 | 415 | IMAEVYRAMKQLDFEW | HHHHHHHHHHHCCCEE | c1a-5 | multi-selected | 0.01 | 0.03 | 0.186 | boundary | boundary|AMPKA2_C; | 0.0 |
| 25 | fp_D | Q09137 | 415 | IMAEVYRAMKQLDFEW | HHHHHHHHHHHCCCEE | c1d-AT-5 | multi | 0.01 | 0.03 | 0.186 | boundary | boundary|AMPKA2_C; | 0.0 |
| 26 | fp_beta_D | Q09137 | 428 | FEWKVVNAYHLRVRR | CEEEEECCEEEEEEE | c1b-AT-4 | multi | 0.01 | 0.023 | 0.24 | boundary | MID|AMPKA2_C; | 0.714 |
| 27 | fp_beta_O | Q09137 | 429 | EWKVVNAYHLRVRR | EEEEECCEEEEEEE | c2-AT-4 | multi-selected | 0.01 | 0.022 | 0.249 | ORD | MID|AMPKA2_C; | 0.714 |
| 28 | fp_beta_D | Q09137 | 443 | KNPVTGNYVKMSLQLYL | ECCCCCCEEEEEEEEEE | c1c-AT-4 | uniq | 0.018 | 0.053 | 0.261 | boundary | MID|AMPKA2_C; | 0.625 |
| 29 | fp_D | Q09137 | 465 | YLLDFKSIDDEVVEQR | EEEEEECCCCHHCCCC | c1aR-4 | uniq | 0.263 | 0.322 | 0.315 | boundary | MID|AMPKA2_C; | 0.286 |
| 30 | cand_D | Q09137 | 528 | ASPRLGSHTMDFFEMCA ++++++++++++++ | CCCCCCCCCCCHHHHHH | c1c-AT-4 | uniq | 0.255 | 0.29 | 0.269 | boundary | boundary|AMPKA2_C; | 0.0 |
| 31 | cand_D | Q09137 | 535 | HTMDFFEMCASLITAL +++++++++++*+++* | CCCCHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.049 | 0.115 | 0.142 | boundary | boundary|AMPKA2_C; | 0.0 |
| 32 | cand_D | Q09137 | 536 | TMDFFEMCASLITA ++++++++++*+++ | CCCHHHHHHHHHHH | c3-AT | multi | 0.041 | 0.104 | 0.138 | boundary | boundary|AMPKA2_C; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment