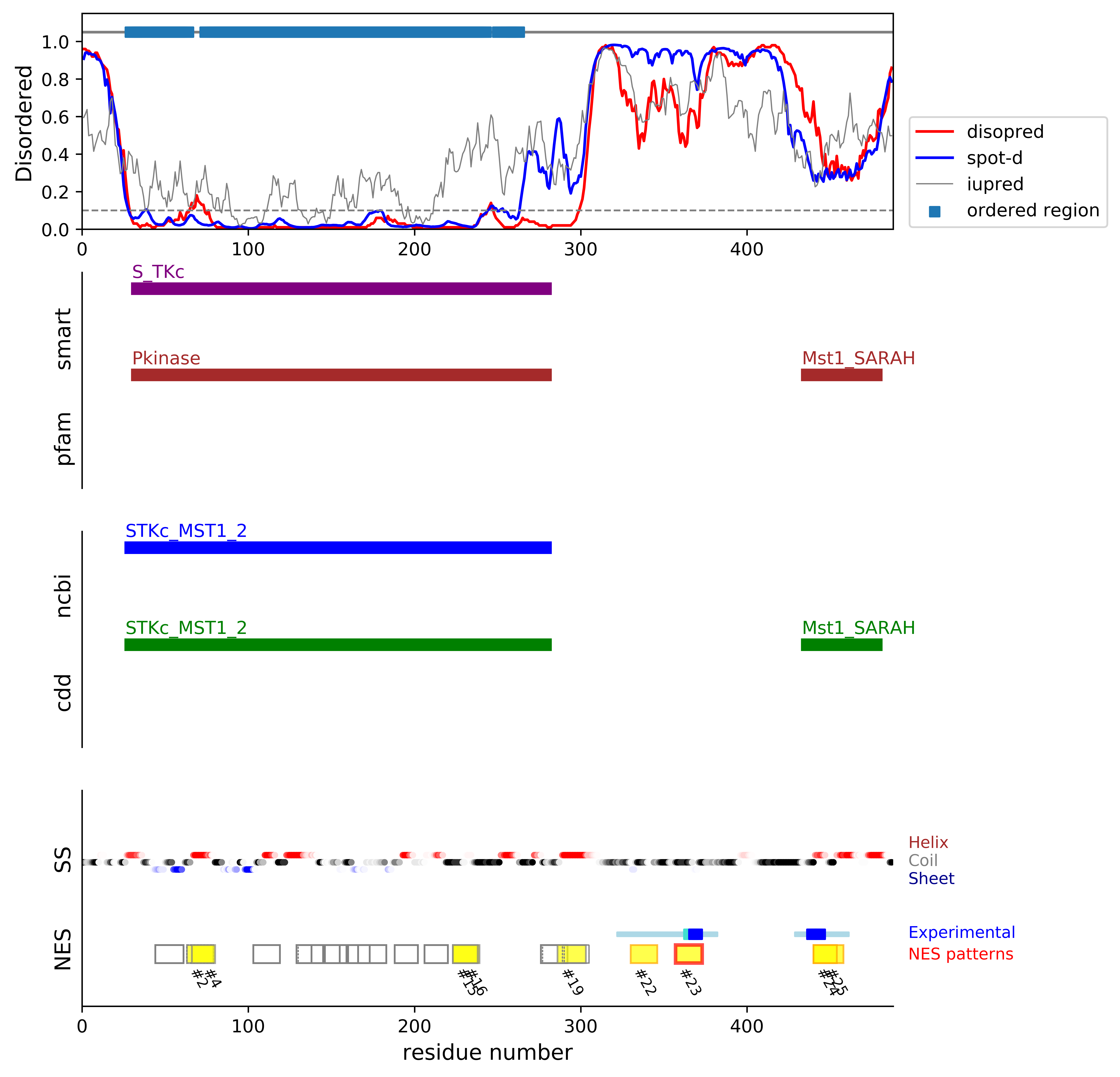

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q13043 | 44 | VYKAIHKETGQIVAIKQ | EEEEEECCCCCEEEEEE | c1c-AT-4 | uniq | 0.039 | 0.033 | 0.229 | boundary | boundary|STKc_MST1_2; | 0.375 |
| 2 | fp_D | Q13043 | 63 | VESDLQEIIKEISIMQ | CCCHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.11 | 0.041 | 0.201 | boundary | MID|STKc_MST1_2; | 0.0 |
| 3 | fp_D | Q13043 | 63 | VESDLQEIIKEISIMQ | CCCHHHHHHHHHHHHH | c1d-4 | multi | 0.11 | 0.041 | 0.201 | boundary | MID|STKc_MST1_2; | 0.0 |
| 4 | fp_D | Q13043 | 66 | DLQEIIKEISIMQQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.107 | 0.036 | 0.207 | boundary | MID|STKc_MST1_2; | 0.0 |
| 5 | fp_O | Q13043 | 103 | EYCGAGSVSDIIRLRN | EECCCCCHHHHHHHCC | c1d-AT-4 | uniq | 0.009 | 0.026 | 0.131 | ORD | MID|STKc_MST1_2; | 0.0 |
| 6 | fp_O | Q13043 | 129 | ILQSTLKGLEYLHFMR | HHHHHHHHHHHHHCCC | c1a-AT-5 | multi-selected | 0.01 | 0.012 | 0.081 | ORD | MID|STKc_MST1_2; | 0.0 |
| 7 | fp_O | Q13043 | 130 | LQSTLKGLEYLHFMR | HHHHHHHHHHHHCCC | c1b-4 | multi | 0.01 | 0.012 | 0.076 | ORD | MID|STKc_MST1_2; | 0.0 |
| 8 | fp_O | Q13043 | 138 | EYLHFMRKIHRDIKAGN | HHHHCCCCCCCCCCCCC | c1cR-4 | uniq | 0.01 | 0.019 | 0.111 | ORD | MID|STKc_MST1_2; | 0.0 |
| 9 | fp_O | Q13043 | 146 | IHRDIKAGNILLNT | CCCCCCCCCEEECC | c2-AT-4 | uniq | 0.01 | 0.022 | 0.156 | ORD | MID|STKc_MST1_2; | 0.143 |
| 10 | fp_beta_O | Q13043 | 159 | TEGHAKLADFGVAG | CCCCEEEEEEEECC | c2-AT-4 | uniq | 0.011 | 0.037 | 0.202 | ORD | MID|STKc_MST1_2; | 0.857 |
| 11 | fp_O | Q13043 | 166 | ADFGVAGQLTDTMAKRN | EEEEECCCCCCCCCCCC | c1cR-4 | uniq | 0.031 | 0.069 | 0.256 | ORD | MID|STKc_MST1_2; | 0.125 |
| 12 | fp_O | Q13043 | 188 | PFWMAPEVIQEIGY | CCCCHHHHHHHHCC | c3-AT | uniq | 0.025 | 0.016 | 0.103 | ORD | MID|STKc_MST1_2; | 0.0 |
| 13 | fp_O | Q13043 | 206 | DIWSLGITAIEMAE | CHHHHHHHHHHHCC | c2-5 | multi-selected | 0.005 | 0.017 | 0.154 | ORD | MID|STKc_MST1_2; | 0.0 |
| 14 | fp_O | Q13043 | 206 | DIWSLGITAIEMAE | CHHHHHHHHHHHCC | c3-AT | multi | 0.005 | 0.017 | 0.154 | ORD | MID|STKc_MST1_2; | 0.0 |
| 15 | fp_D | Q13043 | 223 | PYADIHPMRAIFMIPT | CCCCCCCCHHHHCCCC | c1d-4 | multi-selected | 0.012 | 0.021 | 0.403 | boundary | MID|STKc_MST1_2; | 0.0 |
| 16 | fp_D | Q13043 | 223 | PYADIHPMRAIFMIP | CCCCCCCCHHHHCCC | c1b-4 | multi-selected | 0.011 | 0.02 | 0.393 | boundary | MID|STKc_MST1_2; | 0.0 |
| 17 | fp_D | Q13043 | 276 | LQHPFVRSAKGVSILR | HCCCCCCCCCCCHHHH | c1a-AT-4 | multi-selected | 0.018 | 0.394 | 0.353 | boundary | boundary|STKc_MST1_2; | 0.0 |
| 18 | fp_D | Q13043 | 277 | QHPFVRSAKGVSILR | CCCCCCCCCCCHHHH | c1b-AT-4 | multi | 0.018 | 0.4 | 0.338 | boundary | boundary|STKc_MST1_2; | 0.0 |
| 19 | fp_D | Q13043 | 286 | GVSILRDLINEAMDVKL | CCHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.058 | 0.355 | 0.393 | DISO | boundary|STKc_MST1_2; | 0.0 |
| 20 | fp_D | Q13043 | 289 | ILRDLINEAMDVKLKR | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.104 | 0.341 | 0.431 | DISO | boundary|STKc_MST1_2; | 0.0 |
| 21 | fp_D | Q13043 | 290 | LRDLINEAMDVKLKR | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.11 | 0.332 | 0.437 | DISO | boundary|STKc_MST1_2; | 0.0 |
| 22 | fp_D | Q13043 | 330 | MVRAVGDEMGTVRVAS ................ | CEECCCCCCCCCCCCC | c1a-5 | uniq | 0.619 | 0.93 | 0.645 | DISO | 0.0 | |
| 23 | cand_D | Q13043 | 357 | HDDTLPSQLGTMVINA ........+++*+*.. | CCCCCCCCCCCEEECC | c1a-4 | uniq | 0.569 | 0.898 | 0.716 | DISO | 0.0 | |
| 24 | fp_D | Q13043 | 440 | TVEDLQKRLLALDPMMEQ +*++*............. | CHHHHHHHCCCCCHHHHH | c4-5 | multi-selected | 0.398 | 0.284 | 0.396 | DISO | small|Mst1_SARAH; | 0.0 |
| 25 | fp_D | Q13043 | 440 | TVEDLQKRLLALDP +*++*......... | CHHHHHHHCCCCCH | c3-4 | multi-selected | 0.416 | 0.281 | 0.375 | DISO | small|Mst1_SARAH; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment