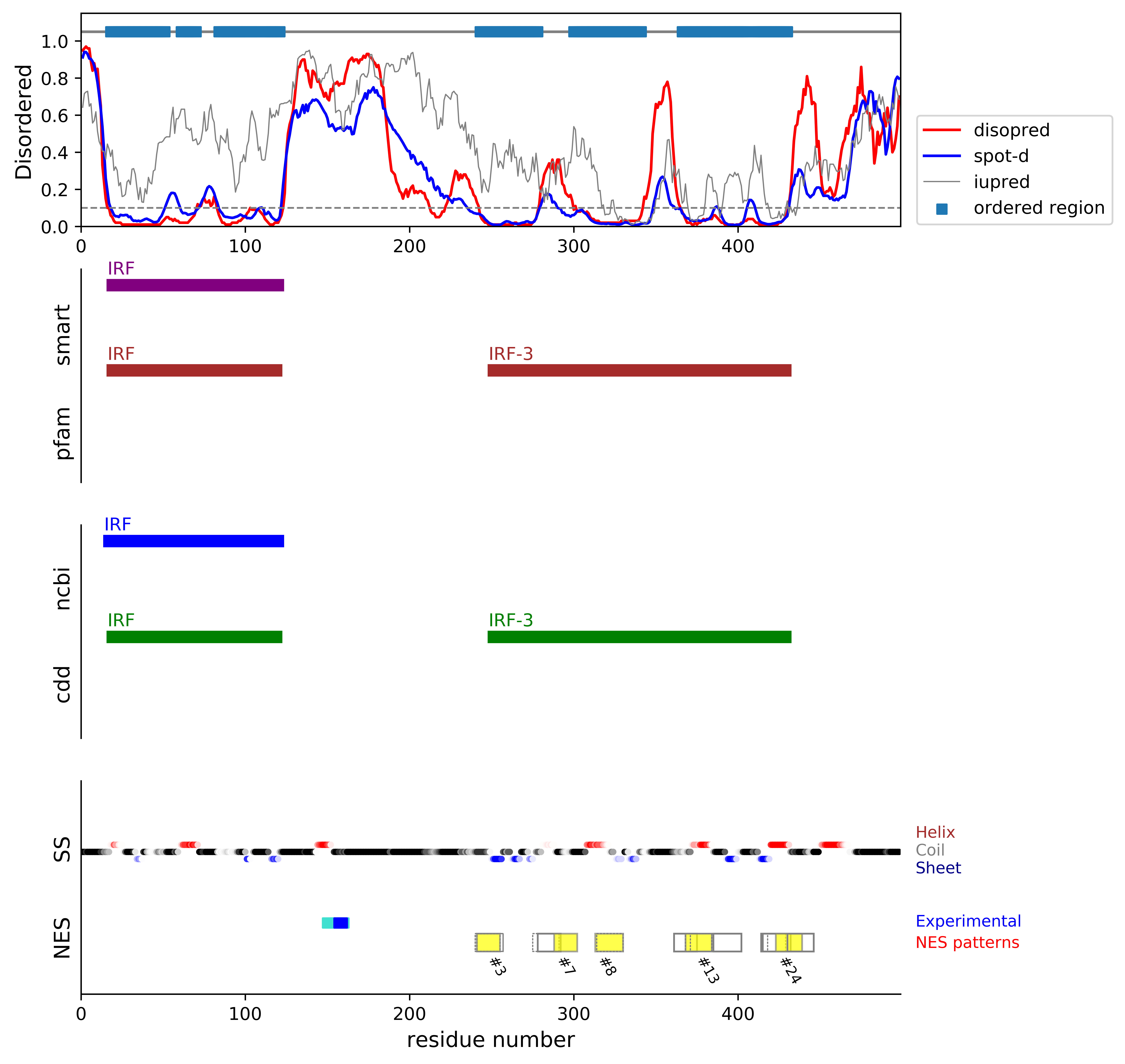

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q13568 | 240 | ISPHMLPLTDLEIKF | CCCCCCCCCCEEEEE | c1b-4 | multi | 0.043 | 0.036 | 0.31 | boundary | boundary|IRF-3; | 0.143 |
| 2 | fp_D | Q13568 | 240 | ISPHMLPLTDLEIKFQY | CCCCCCCCCCEEEEEEE | c1c-AT-4 | multi | 0.039 | 0.033 | 0.315 | boundary | boundary|IRF-3; | 0.25 |
| 3 | fp_D | Q13568 | 241 | SPHMLPLTDLEIKF | CCCCCCCCCEEEEE | c2-4 | multi-selected | 0.034 | 0.033 | 0.305 | boundary | boundary|IRF-3; | 0.143 |
| 4 | fp_D | Q13568 | 241 | SPHMLPLTDLEIKFQY | CCCCCCCCCEEEEEEE | c1d-AT-4 | multi | 0.031 | 0.03 | 0.31 | boundary | boundary|IRF-3; | 0.286 |
| 5 | fp_D | Q13568 | 275 | FYSQLEATQEQVELFG | ECCCCCHHHHHHHHCC | c1d-AT-4 | multi | 0.226 | 0.116 | 0.266 | boundary | MID|IRF-3; | 0.0 |
| 6 | fp_D | Q13568 | 278 | QLEATQEQVELFGP | CCCHHHHHHHHCCC | c3-AT | multi-selected | 0.277 | 0.133 | 0.25 | boundary | MID|IRF-3; | 0.0 |
| 7 | fp_D | Q13568 | 288 | LFGPISLEQVRFPS | HCCCCCCCCCCCCC | c2-5 | uniq | 0.221 | 0.08 | 0.358 | boundary | MID|IRF-3; | 0.0 |
| 8 | fp_D | Q13568 | 313 | YTNQLLDVLDRGLILQL | HHHHHHHHHCCCEEEEE | c1c-4 | multi-selected | 0.022 | 0.014 | 0.119 | boundary | MID|IRF-3; | 0.0 |
| 9 | fp_D | Q13568 | 314 | TNQLLDVLDRGLILQL | HHHHHHHHCCCEEEEE | c1d-4 | multi | 0.022 | 0.014 | 0.108 | boundary | MID|IRF-3; | 0.0 |
| 10 | fp_D | Q13568 | 361 | IQREVKTKLFSLEH | CCCCCCCCCCCHHH | c2-AT-4 | uniq | 0.089 | 0.064 | 0.16 | boundary | MID|IRF-3; | 0.0 |
| 11 | fp_D | Q13568 | 368 | KLFSLEHFLNELILFQK | CCCCHHHHHHHHHHHHH | c1c-5 | multi | 0.026 | 0.037 | 0.119 | boundary | MID|IRF-3; | 0.0 |
| 12 | fp_D | Q13568 | 368 | KLFSLEHFLNELILFQK | CCCCHHHHHHHHHHHHH | c1cR-4 | multi | 0.026 | 0.037 | 0.119 | boundary | MID|IRF-3; | 0.0 |
| 13 | fp_D | Q13568 | 368 | KLFSLEHFLNELILFQ | CCCCHHHHHHHHHHHH | c1a-5 | multi-selected | 0.026 | 0.036 | 0.11 | boundary | MID|IRF-3; | 0.0 |
| 14 | fp_D | Q13568 | 368 | KLFSLEHFLNELILFQ | CCCCHHHHHHHHHHHH | c1aR-5 | multi | 0.026 | 0.036 | 0.11 | boundary | MID|IRF-3; | 0.0 |
| 15 | fp_D | Q13568 | 368 | KLFSLEHFLNELILFQ | CCCCHHHHHHHHHHHH | c1d-5 | multi | 0.026 | 0.036 | 0.11 | boundary | MID|IRF-3; | 0.0 |
| 16 | fp_D | Q13568 | 371 | SLEHFLNELILFQK | CHHHHHHHHHHHHH | c3-4 | multi | 0.026 | 0.036 | 0.114 | boundary | MID|IRF-3; | 0.0 |
| 17 | fp_beta_D | Q13568 | 385 | GQTNTPPPFEIFFCFGE | CCCCCCCCEEEEEEECC | c1c-AT-4 | uniq | 0.022 | 0.039 | 0.212 | boundary | MID|IRF-3; | 0.5 |
| 18 | fp_D | Q13568 | 414 | ITVQVVPVAARLLLEM | EEEEEEHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.017 | 0.021 | 0.108 | boundary | boundary|IRF-3; | 0.286 |
| 19 | fp_D | Q13568 | 414 | ITVQVVPVAARLLLEM | EEEEEEHHHHHHHHHH | c1d-4 | multi | 0.017 | 0.021 | 0.108 | boundary | boundary|IRF-3; | 0.286 |
| 20 | fp_D | Q13568 | 415 | TVQVVPVAARLLLEMFS | EEEEEHHHHHHHHHHHH | c1c-AT-5 | multi-selected | 0.027 | 0.028 | 0.092 | boundary | boundary|IRF-3; | 0.125 |
| 21 | fp_D | Q13568 | 415 | TVQVVPVAARLLLEM | EEEEEHHHHHHHHHH | c1b-AT-5 | multi | 0.017 | 0.021 | 0.09 | boundary | boundary|IRF-3; | 0.143 |
| 22 | fp_D | Q13568 | 415 | TVQVVPVAARLLLE | EEEEEHHHHHHHHH | c3-AT | multi | 0.015 | 0.019 | 0.09 | boundary | boundary|IRF-3; | 0.286 |
| 23 | fp_D | Q13568 | 418 | VVPVAARLLLEMFS | EEHHHHHHHHHHHH | c3-AT | multi | 0.031 | 0.031 | 0.071 | boundary | boundary|IRF-3; | 0.0 |
| 24 | fp_D | Q13568 | 423 | ARLLLEMFSGELSWSA | HHHHHHHHHCCCCCCC | c1aR-4 | multi-selected | 0.229 | 0.133 | 0.094 | boundary | boundary|IRF-3; | 0.0 |
| 25 | fp_D | Q13568 | 430 | FSGELSWSADSIRLQI | HHCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.534 | 0.21 | 0.199 | boundary | boundary|IRF-3; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment