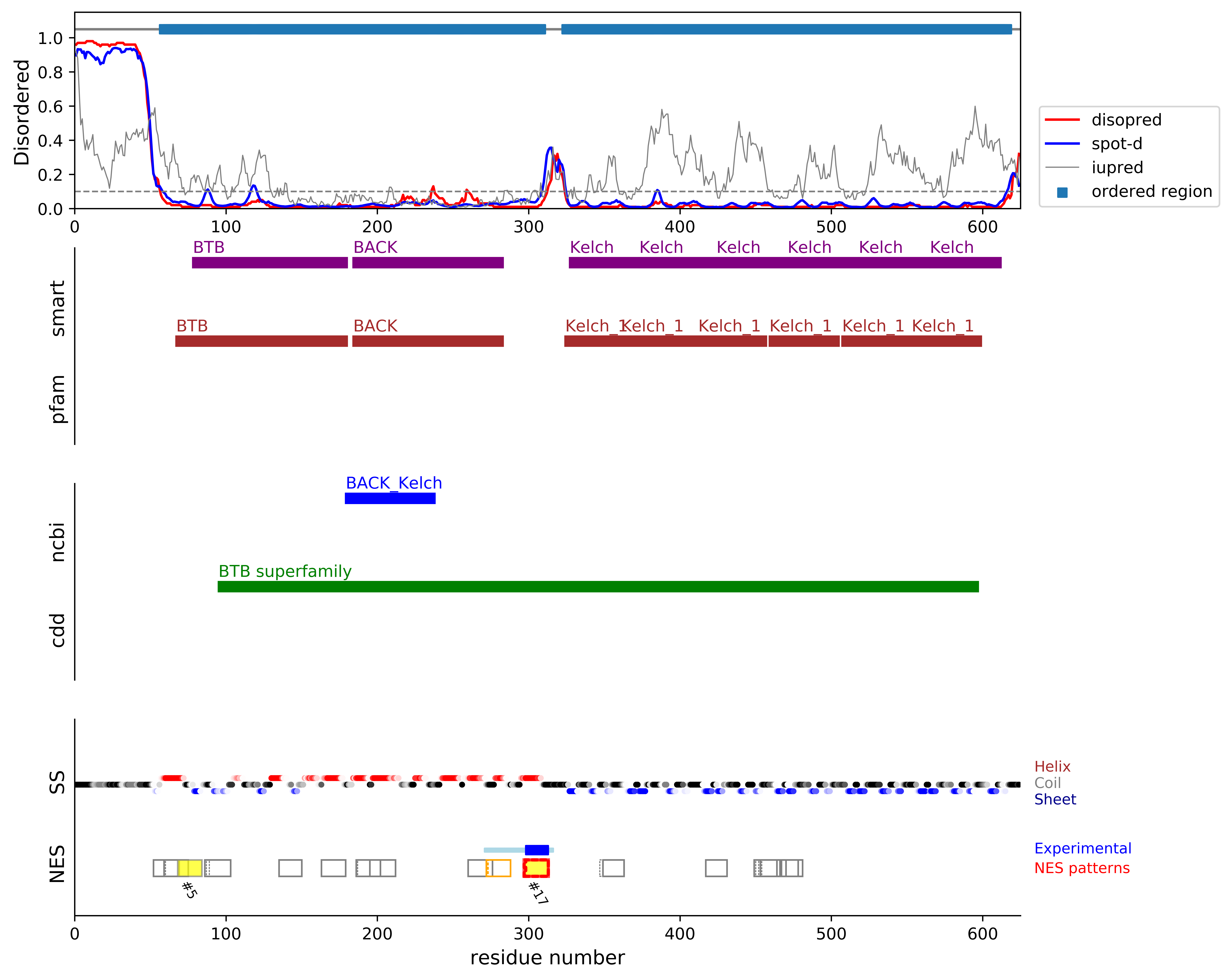

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14145 | 52 | FSYTLEDHTKQAFGIMN | CEEECHHHHHHHHHHHH | c1c-AT-4 | uniq | 0.075 | 0.092 | 0.337 | boundary | 0.0 | |
| 2 | fp_D | Q14145 | 59 | HTKQAFGIMNELRLSQ | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi-selected | 0.023 | 0.049 | 0.22 | boundary | boundary|BTB superfamily; | 0.0 |
| 3 | fp_D | Q14145 | 59 | HTKQAFGIMNELRLSQ | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.023 | 0.049 | 0.22 | boundary | boundary|BTB superfamily; | 0.0 |
| 4 | fp_D | Q14145 | 60 | TKQAFGIMNELRLSQ | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.021 | 0.046 | 0.212 | boundary | boundary|BTB superfamily; | 0.0 |
| 5 | fp_D | Q14145 | 68 | NELRLSQQLCDVTLQV | HHHHHCCCCCCEEEEE | c1a-4 | uniq | 0.013 | 0.028 | 0.156 | boundary | boundary|BTB superfamily; | 0.0 |
| 6 | fp_beta_D | Q14145 | 86 | QDAPAAQFMAHKVVLAS | CCCCCCEEEEEEEEEEC | c1c-AT-4 | multi-selected | 0.013 | 0.044 | 0.142 | boundary | boundary|BTB superfamily; | 0.75 |
| 7 | fp_beta_D | Q14145 | 87 | DAPAAQFMAHKVVLAS | CCCCCEEEEEEEEEEC | c1d-AT-4 | multi | 0.013 | 0.041 | 0.144 | boundary | boundary|BTB superfamily; | 0.857 |
| 8 | fp_beta_D | Q14145 | 89 | PAAQFMAHKVVLAS | CCCEEEEEEEEEEC | c2-AT-4 | multi | 0.011 | 0.032 | 0.139 | boundary | boundary|BTB superfamily; | 1.0 |
| 9 | fp_O | Q14145 | 135 | RLIEFAYTASISMGE | HHHHHHCCCEEEECC | c1b-AT-5 | uniq | 0.01 | 0.017 | 0.069 | ORD | MID|BTB superfamily; | 0.286 |
| 10 | fp_O | Q14145 | 163 | QIDSVVRACSDFLVQQ | CHHHHHHHHHHHHHHC | c1d-AT-5 | uniq | 0.014 | 0.012 | 0.041 | ORD | MID|BTB superfamily; | 0.0 |
| 11 | fp_O | Q14145 | 186 | GIANFAEQIGCVELHQ | HHHHHHHHHCCHHHHH | c1a-5 | multi-selected | 0.01 | 0.02 | 0.056 | ORD | MID|BTB superfamily; | 0.0 |
| 12 | fp_O | Q14145 | 187 | IANFAEQIGCVELHQ | HHHHHHHHCCHHHHH | c1b-AT-4 | multi | 0.01 | 0.021 | 0.054 | ORD | MID|BTB superfamily; | 0.0 |
| 13 | fp_O | Q14145 | 195 | GCVELHQRAREYIYMHF | CCHHHHHHHHHHHHHHH | c1c-AT-4 | uniq | 0.011 | 0.02 | 0.057 | ORD | MID|BTB superfamily; | 0.0 |
| 14 | fp_O | Q14145 | 260 | RRFYVQALLRAVRCHS .... | HHHHHHHHHHHHCCCC | c1aR-4 | uniq | 0.046 | 0.021 | 0.026 | ORD | MID|BTB superfamily; | 0.0 |
| 15 | fp_O | Q14145 | 272 | RCHSLTPNFLQMQLQK ................ | CCCCCCHHHHHHHHCC | c1a-4 | multi-selected | 0.014 | 0.027 | 0.054 | ORD | MID|BTB superfamily; | 0.0 |
| 16 | fp_O | Q14145 | 273 | CHSLTPNFLQMQLQK ............... | CCCCCHHHHHHHHCC | c1b-AT-4 | multi | 0.013 | 0.028 | 0.057 | ORD | MID|BTB superfamily; | 0.0 |
| 17 | cand_D | Q14145 | 297 | CKDYLVKIFEELTLHK ....*..*...*.*.. | HHHHHHHHHHHHCCCC | c1a-4 | multi-selected | 0.024 | 0.087 | 0.081 | boundary | MID|BTB superfamily; | 0.0 |
| 18 | cand_D | Q14145 | 297 | CKDYLVKIFEELTLHK ....*..*...*.*.. | HHHHHHHHHHHHCCCC | c1d-4 | multi | 0.024 | 0.087 | 0.081 | boundary | MID|BTB superfamily; | 0.0 |
| 19 | cand_D | Q14145 | 298 | KDYLVKIFEELTLHK ...*..*...*.*.. | HHHHHHHHHHHCCCC | c1b-4 | multi | 0.025 | 0.089 | 0.084 | boundary | MID|BTB superfamily; | 0.0 |
| 20 | fp_O | Q14145 | 347 | PSDGTWLRLADLQVPR | CCCCCEEECCCCCCCC | c1a-AT-4 | multi | 0.011 | 0.033 | 0.181 | ORD | MID|BTB superfamily; | 0.429 |
| 21 | fp_O | Q14145 | 349 | DGTWLRLADLQVPR | CCCEEECCCCCCCC | c2-4 | multi-selected | 0.011 | 0.035 | 0.188 | ORD | MID|BTB superfamily; | 0.429 |
| 22 | fp_beta_O | Q14145 | 417 | GVGVIDGHIYAVGG | EEEEECCCEEEECC | c3-4 | uniq | 0.009 | 0.009 | 0.138 | ORD | MID|BTB superfamily; | 0.571 |
| 23 | fp_O | Q14145 | 449 | EWHLVAPMLTRRIGVGV | EEEEECCCCCCCCCCEE | c1c-4 | multi-selected | 0.019 | 0.025 | 0.205 | ORD | MID|BTB superfamily; | 0.125 |
| 24 | fp_O | Q14145 | 450 | WHLVAPMLTRRIGVGV | EEEECCCCCCCCCCEE | c1d-AT-4 | multi | 0.019 | 0.024 | 0.194 | ORD | MID|BTB superfamily; | 0.0 |
| 25 | fp_O | Q14145 | 452 | LVAPMLTRRIGVGV | EECCCCCCCCCCEE | c2-AT-5 | multi | 0.018 | 0.023 | 0.169 | ORD | MID|BTB superfamily; | 0.0 |
| 26 | fp_O | Q14145 | 453 | VAPMLTRRIGVGVAVLN | ECCCCCCCCCCEEEEEC | c1c-4 | multi-selected | 0.015 | 0.019 | 0.135 | ORD | MID|BTB superfamily; | 0.125 |
| 27 | fp_O | Q14145 | 454 | APMLTRRIGVGVAVLN | CCCCCCCCCCEEEEEC | c1d-AT-4 | multi | 0.014 | 0.019 | 0.126 | ORD | MID|BTB superfamily; | 0.143 |
| 28 | fp_beta_O | Q14145 | 464 | GVAVLNRLLYAVGG | EEEEECCCEEEECC | c3-4 | multi-selected | 0.009 | 0.01 | 0.087 | ORD | MID|BTB superfamily; | 0.571 |
| 29 | fp_beta_O | Q14145 | 467 | VLNRLLYAVGGFDG | EECCCEEEECCCCC | c3-4 | multi-selected | 0.009 | 0.017 | 0.106 | ORD | MID|BTB superfamily; | 0.571 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment