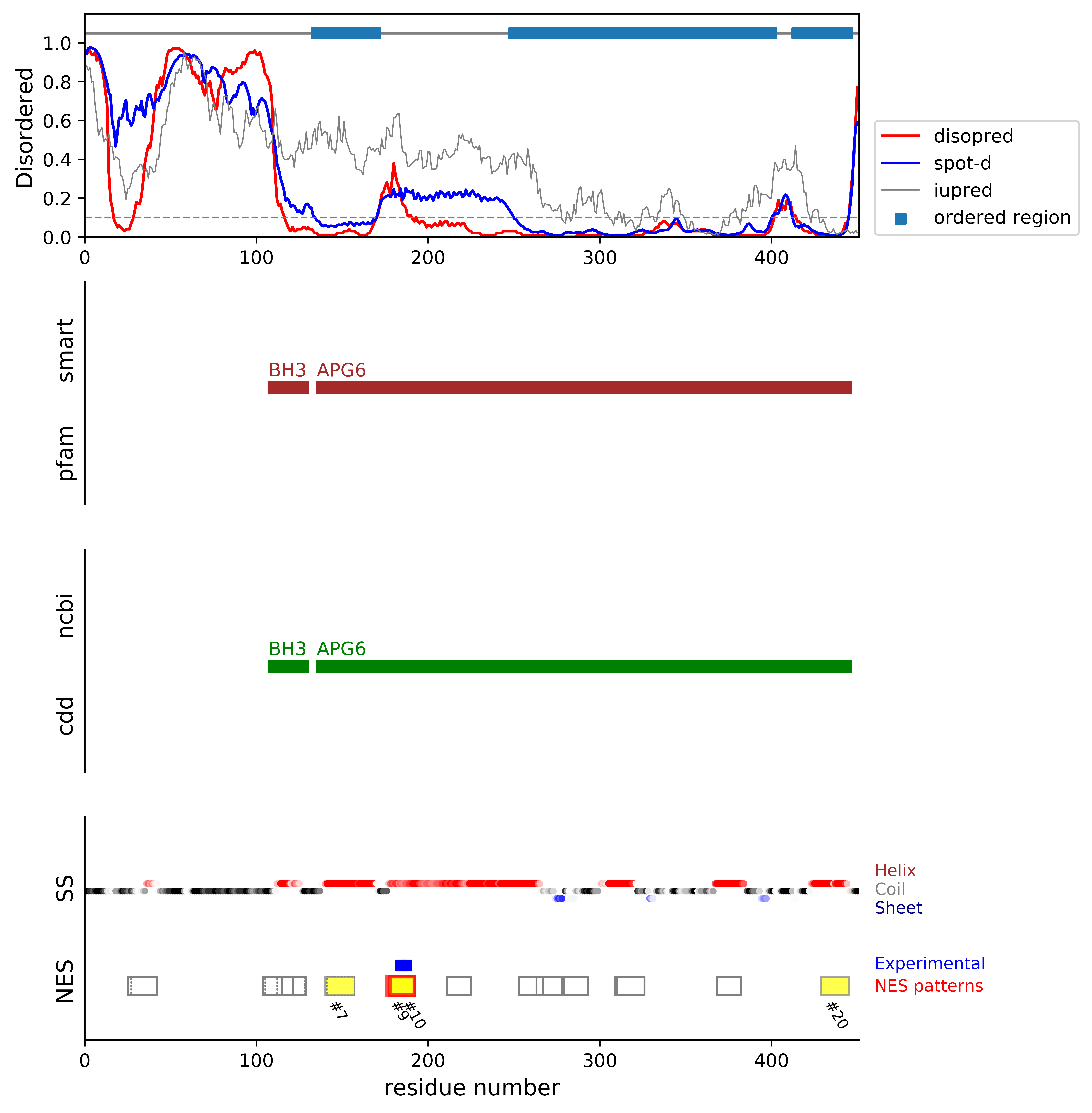

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14457 | 25 | LKLDTSFKILDRVTIQE | CCCCCCCCCCCHHHHHH | c1c-AT-4 | multi-selected | 0.293 | 0.656 | 0.33 | DISO | 0.0 | |
| 2 | fp_D | Q14457 | 27 | LDTSFKILDRVTIQE | CCCCCCCCCHHHHHH | c1b-4 | multi | 0.327 | 0.664 | 0.341 | DISO | 0.0 | |
| 3 | fp_D | Q14457 | 104 | SDGGTMENLSRRLKVTG | CCCCCCCCHHHHHHHHH | c1c-AT-4 | multi-selected | 0.387 | 0.416 | 0.482 | boundary | boundary|APG6; | 0.0 |
| 4 | fp_D | Q14457 | 105 | DGGTMENLSRRLKVTG | CCCCCCCHHHHHHHHH | c1d-4 | multi | 0.356 | 0.398 | 0.478 | boundary | boundary|APG6; | 0.0 |
| 5 | fp_D | Q14457 | 112 | LSRRLKVTGDLFDIMS | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.078 | 0.203 | 0.439 | boundary | small|BH3; boundary|APG6; | 0.0 |
| 6 | fp_D | Q14457 | 115 | RLKVTGDLFDIMSG | HHHHHHHHHHHCCC | c3-AT | multi-selected | 0.056 | 0.169 | 0.425 | boundary | boundary|APG6; | 0.0 |
| 7 | fp_D | Q14457 | 140 | CTDTLLDQLDTQLNVTE | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.02 | 0.062 | 0.494 | boundary | boundary|APG6; | 0.0 |
| 8 | fp_D | Q14457 | 141 | TDTLLDQLDTQLNVTE | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.021 | 0.062 | 0.491 | __ | boundary|APG6; | 0.0 |
| 9 | cand_D | Q14457 | 176 | DSEQLQMELKELALEE * * | CHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.212 | 0.223 | 0.499 | boundary | MID|APG6; | 0.0 |
| 10 | cand_D | Q14457 | 178 | EQLQMELKELALEE * * | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.205 | 0.224 | 0.49 | boundary | MID|APG6; | 0.0 |
| 11 | fp_D | Q14457 | 211 | NLEKVQAEAERLDQ | HHHHHHHHHHHHHH | c3-AT | uniq | 0.061 | 0.22 | 0.466 | DISO | MID|APG6; | 0.0 |
| 12 | fp_D | Q14457 | 253 | QMRYAQTQLDKLKK | HHHHHHHHHHHHHC | c3-AT | uniq | 0.011 | 0.037 | 0.343 | boundary | MID|APG6; | 0.0 |
| 13 | fp_D | Q14457 | 263 | KLKKTNVFNATFHIWH | HHHCCCCCCCCEEEEE | c1d-AT-5 | uniq | 0.007 | 0.017 | 0.172 | __ | MID|APG6; | 0.0 |
| 14 | fp_O | Q14457 | 278 | HSGQFGTINNFRLGR | ECCCCEEECCCCCCC | c1b-4 | uniq | 0.004 | 0.021 | 0.145 | ORD | MID|APG6; | 0.429 |
| 15 | fp_O | Q14457 | 309 | QTVLLLHALANKMGLKF | HHHHHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.009 | 0.017 | 0.084 | ORD | MID|APG6; | 0.0 |
| 16 | fp_O | Q14457 | 310 | TVLLLHALANKMGLKF | HHHHHHHHHHHHCCCC | c1aR-4 | multi-selected | 0.01 | 0.018 | 0.083 | ORD | MID|APG6; | 0.0 |
| 17 | fp_O | Q14457 | 310 | TVLLLHALANKMGLKF | HHHHHHHHHHHHCCCC | c1a-AT-5 | multi | 0.01 | 0.018 | 0.083 | ORD | MID|APG6; | 0.0 |
| 18 | fp_O | Q14457 | 310 | TVLLLHALANKMGLKF | HHHHHHHHHHHHCCCC | c1d-5 | multi | 0.01 | 0.018 | 0.083 | ORD | MID|APG6; | 0.0 |
| 19 | fp_O | Q14457 | 368 | AMVAFLDCVQQFKE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.009 | 0.013 | 0.077 | ORD | MID|APG6; | 0.0 |
| 20 | fp_D | Q14457 | 429 | LKFMLTNLKWGLAWVS | HHHHHHHHHHHHHHHH | c1aR-5 | uniq | 0.018 | 0.016 | 0.037 | boundary | boundary|APG6; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment