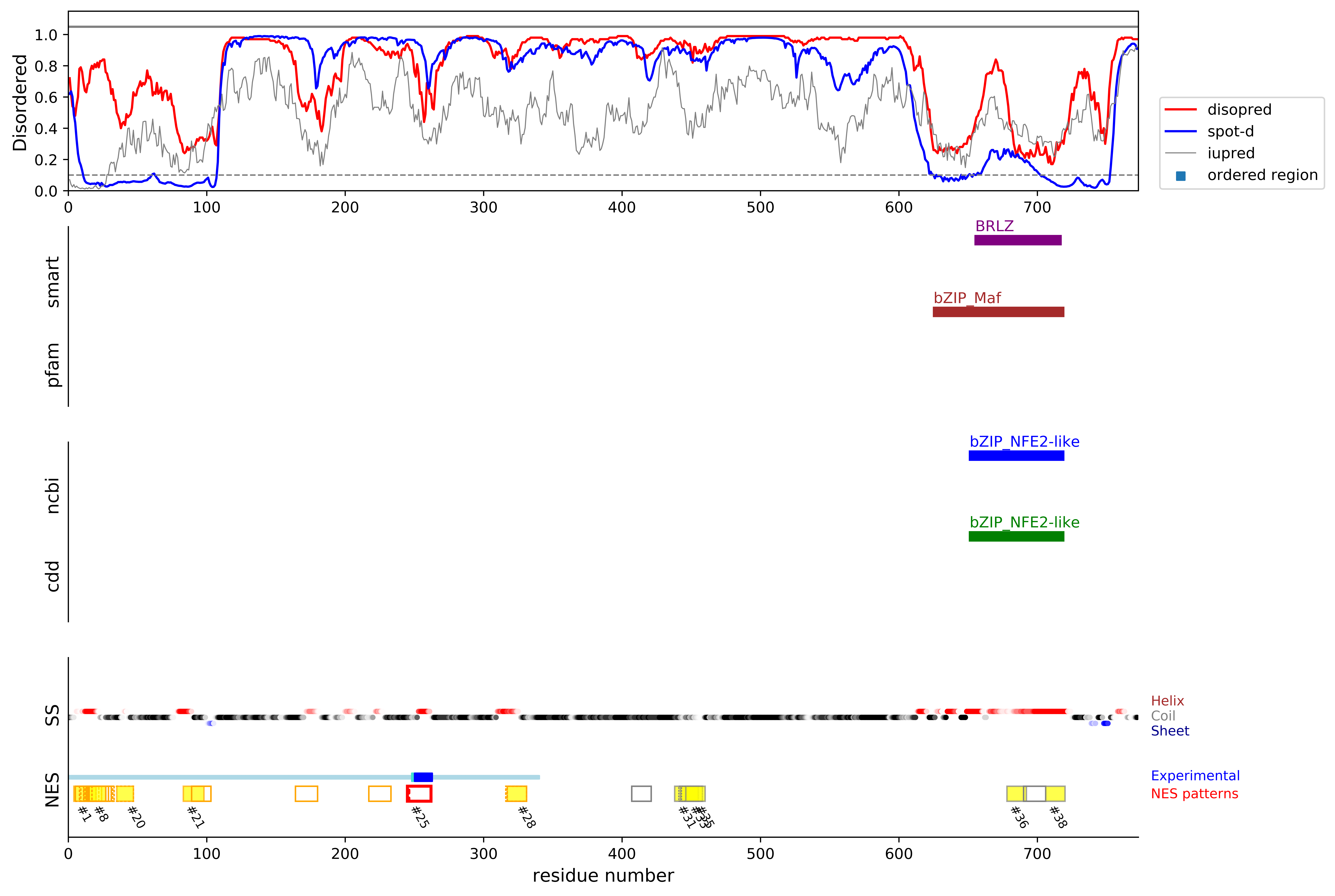

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14494 | 4 | LKKYLTEGLLQFTILL ................ | CHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.686 | 0.153 | 0.019 | DISO | 0.0 | |
| 2 | fp_D | Q14494 | 5 | KKYLTEGLLQFTILL ............... | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.697 | 0.13 | 0.018 | DISO | 0.0 | |
| 3 | fp_D | Q14494 | 5 | KKYLTEGLLQFTILLSL ................. | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.709 | 0.12 | 0.018 | DISO | 0.0 | |
| 4 | fp_D | Q14494 | 8 | LTEGLLQFTILLSLIG ................ | HHHHHHHHHHHHHHHC | c1a-AT-4 | multi | 0.753 | 0.07 | 0.017 | DISO | 0.0 | |
| 5 | fp_D | Q14494 | 8 | LTEGLLQFTILLSLIG ................ | HHHHHHHHHHHHHHHC | c1d-4 | multi | 0.753 | 0.07 | 0.017 | DISO | 0.0 | |
| 6 | fp_D | Q14494 | 9 | TEGLLQFTILLSLIG ............... | HHHHHHHHHHHHHHC | c1b-AT-4 | multi | 0.751 | 0.063 | 0.017 | DISO | 0.0 | |
| 7 | fp_D | Q14494 | 11 | GLLQFTILLSLIGVRV ................ | HHHHHHHHHHHHCCCC | c1aR-4 | multi | 0.764 | 0.048 | 0.02 | DISO | 0.0 | |
| 8 | fp_D | Q14494 | 11 | GLLQFTILLSLIGVRV ................ | HHHHHHHHHHHHCCCC | c1a-5 | multi-selected | 0.764 | 0.048 | 0.02 | DISO | 0.0 | |
| 9 | fp_D | Q14494 | 11 | GLLQFTILLSLIGVRV ................ | HHHHHHHHHHHHCCCC | c1d-5 | multi | 0.764 | 0.048 | 0.02 | DISO | 0.0 | |
| 10 | fp_D | Q14494 | 12 | LLQFTILLSLIGVRV ............... | HHHHHHHHHHHCCCC | c1b-AT-5 | multi | 0.769 | 0.045 | 0.021 | DISO | 0.0 | |
| 11 | fp_D | Q14494 | 13 | LQFTILLSLIGVRVDV ................ | HHHHHHHHHHCCCCCC | c1a-4 | multi | 0.776 | 0.042 | 0.028 | DISO | 0.0 | |
| 12 | fp_D | Q14494 | 13 | LQFTILLSLIGVRV .............. | HHHHHHHHHHCCCC | c2-4 | multi | 0.779 | 0.044 | 0.021 | DISO | 0.0 | |
| 13 | fp_D | Q14494 | 14 | QFTILLSLIGVRVDVDT ................. | HHHHHHHHHCCCCCCCC | c1c-5 | multi | 0.776 | 0.04 | 0.041 | DISO | 0.0 | |
| 14 | fp_D | Q14494 | 14 | QFTILLSLIGVRVDV ............... | HHHHHHHHHCCCCCC | c1b-5 | multi | 0.785 | 0.041 | 0.029 | DISO | 0.0 | |
| 15 | fp_D | Q14494 | 15 | FTILLSLIGVRVDVDT ................ | HHHHHHHHCCCCCCCC | c1aR-4 | multi | 0.782 | 0.04 | 0.043 | DISO | 0.0 | |
| 16 | fp_D | Q14494 | 15 | FTILLSLIGVRVDVDT ................ | HHHHHHHHCCCCCCCC | c1d-4 | multi | 0.782 | 0.04 | 0.043 | DISO | 0.0 | |
| 17 | fp_D | Q14494 | 15 | FTILLSLIGVRVDV .............. | HHHHHHHHCCCCCC | c2-4 | multi | 0.793 | 0.041 | 0.03 | DISO | 0.0 | |
| 18 | fp_D | Q14494 | 17 | ILLSLIGVRVDVDTYL ................ | HHHHHHCCCCCCCCCC | c1aR-4 | multi | 0.77 | 0.039 | 0.063 | DISO | 0.0 | |
| 19 | fp_D | Q14494 | 32 | LTSQLPPLREIILGP ............... | CCCCCCCCHHHCCCC | c1b-4 | multi | 0.493 | 0.052 | 0.256 | DISO | 0.0 | |
| 20 | fp_D | Q14494 | 35 | QLPPLREIILGP ............ | CCCCCHHHCCCC | c2-rev | multi-selected | 0.465 | 0.055 | 0.28 | DISO | 0.0 | |
| 21 | fp_D | Q14494 | 83 | LLSQVRALDRFQVPT ............... | HHHHHHHCCCCCCCC | c1b-5 | uniq | 0.291 | 0.038 | 0.193 | DISO | 0.0 | |
| 22 | fp_D | Q14494 | 89 | ALDRFQVPTTEVNA .............. | HCCCCCCCCCCEEE | c3-AT | uniq | 0.319 | 0.053 | 0.273 | DISO | 0.0 | |
| 23 | fp_D | Q14494 | 164 | SGDLTKEDIDLIDILW ................ | CCCCCCCHHHHHHHHH | c1a-AT-4 | uniq | 0.593 | 0.924 | 0.358 | DISO | 0.0 | |
| 24 | fp_D | Q14494 | 217 | AGEGAEALARNLLVDG ................ | CCCCHHHHHHHCCCCC | c1d-AT-4 | uniq | 0.908 | 0.975 | 0.563 | DISO | 0.0 | |
| 25 | cand_D | Q14494 | 245 | GEDQTALSLEECLRLLE ......++*+++*++*. | CCCCCCCCHHHHHHHHH | c1c-AT-4 | multi-selected | 0.722 | 0.89 | 0.457 | DISO | 0.0 | |
| 26 | cand_D | Q14494 | 246 | EDQTALSLEECLRLLE .....++*+++*++*. | CCCCCCCHHHHHHHHH | c1d-AT-4 | multi | 0.709 | 0.885 | 0.438 | DISO | 0.0 | |
| 27 | fp_D | Q14494 | 316 | DLMSIMEMQAMEVNT ............... | HHHHHHHHHHCCCCC | c1b-5 | multi | 0.881 | 0.816 | 0.423 | DISO | 0.0 | |
| 28 | fp_D | Q14494 | 317 | LMSIMEMQAMEVNT .............. | HHHHHHHHHCCCCC | c2-5 | multi-selected | 0.879 | 0.813 | 0.423 | DISO | 0.0 | |
| 29 | fp_D | Q14494 | 317 | LMSIMEMQAMEVNT .............. | HHHHHHHHHCCCCC | c3-AT | multi | 0.879 | 0.813 | 0.423 | DISO | 0.0 | |
| 30 | fp_D | Q14494 | 407 | TFGSTNLTGLFFPP | CCCCCCCCCCCCCC | c2-AT-5 | uniq | 0.882 | 0.867 | 0.422 | DISO | 0.0 | |
| 31 | fp_D | Q14494 | 438 | LGGLLDEAMLDEISLMD | CCCCCCCCCCCCCCCCC | c1c-4 | multi-selected | 0.925 | 0.88 | 0.518 | DISO | 0.0 | |
| 32 | fp_D | Q14494 | 441 | LLDEAMLDEISLMD | CCCCCCCCCCCCCC | c2-AT-5 | multi | 0.92 | 0.872 | 0.473 | DISO | 0.0 | |
| 33 | fp_D | Q14494 | 442 | LDEAMLDEISLMDLAI | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.913 | 0.863 | 0.448 | DISO | 0.0 | |
| 34 | fp_D | Q14494 | 443 | DEAMLDEISLMDLAI | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.91 | 0.861 | 0.437 | DISO | 0.0 | |
| 35 | fp_D | Q14494 | 446 | MLDEISLMDLAIEE | CCCCCCCCCCCCCC | c2-5 | multi-selected | 0.904 | 0.85 | 0.42 | DISO | 0.0 | |
| 36 | fp_D | Q14494 | 678 | TILNLERDVEDLQR | HHHHHHHHHHHHHH | c3-4 | uniq | 0.331 | 0.224 | 0.4 | DISO | MID|bZIP_NFE2-like; | 0.0 |
| 37 | fp_D | Q14494 | 690 | QRDKARLLREKVEFLR | HHHHHHHHHHHHHHHH | c1d-AT-4 | uniq | 0.239 | 0.142 | 0.341 | DISO | boundary|bZIP_NFE2-like; | 0.0 |
| 38 | fp_D | Q14494 | 706 | SLRQMKQKVQSLYQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.247 | 0.049 | 0.306 | DISO | boundary|bZIP_NFE2-like; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment