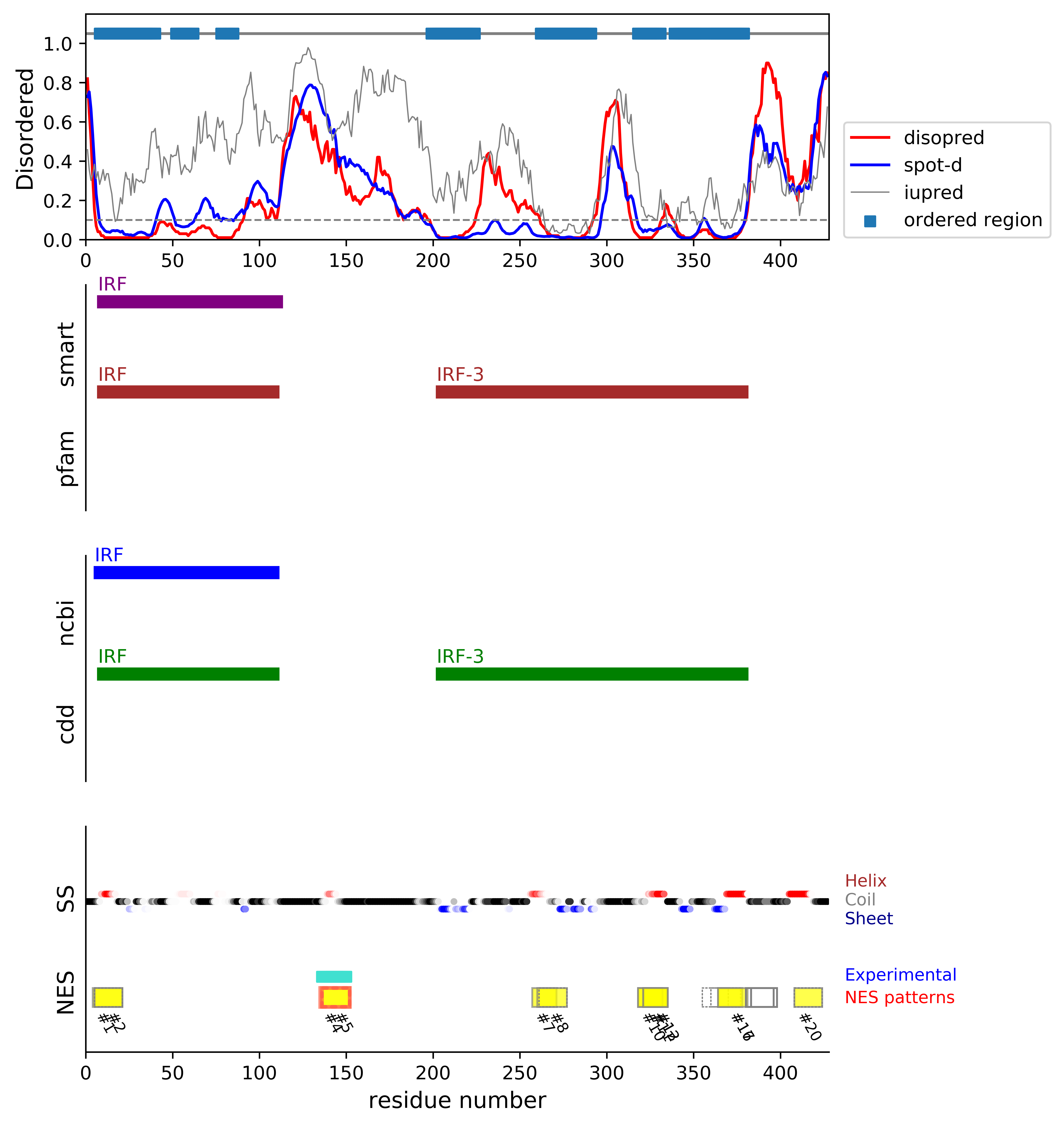

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14653 | 4 | PKPRILPWLVSQLDLGQ | CCCCCHHHHHHHHHCCC | c1c-4 | multi-selected | 0.045 | 0.115 | 0.266 | boundary | boundary|IRF; | 0.0 |
| 2 | fp_D | Q14653 | 5 | KPRILPWLVSQLDLGQ | CCCCHHHHHHHHHCCC | c1a-4 | multi-selected | 0.028 | 0.088 | 0.262 | boundary | boundary|IRF; | 0.0 |
| 3 | fp_D | Q14653 | 5 | KPRILPWLVSQLDLGQ | CCCCHHHHHHHHHCCC | c1d-4 | multi | 0.028 | 0.088 | 0.262 | boundary | boundary|IRF; | 0.0 |
| 4 | cand_D | Q14653 | 135 | TQEDILDELLGNMVLAP +++++++++++++++ | CCCCHHHHHHCCCCCCC | c1c-4 | multi-selected | 0.385 | 0.52 | 0.616 | DISO | boundary|IRF; | 0.0 |
| 5 | cand_D | Q14653 | 136 | QEDILDELLGNMVLAP +++++++++++++++ | CCCHHHHHHCCCCCCC | c1a-4 | multi-selected | 0.382 | 0.509 | 0.605 | DISO | boundary|IRF; | 0.0 |
| 6 | cand_D | Q14653 | 136 | QEDILDELLGNMVLAP +++++++++++++++ | CCCHHHHHHCCCCCCC | c1d-4 | multi | 0.382 | 0.509 | 0.605 | DISO | boundary|IRF; | 0.0 |
| 7 | fp_D | Q14653 | 257 | VMSYVRHVLSCLGG | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.075 | 0.02 | 0.11 | boundary | MID|IRF-3; | 0.0 |
| 8 | fp_D | Q14653 | 260 | YVRHVLSCLGGGLALWR | HHHHHHHCCCCCCEEEE | c1c-5 | multi-selected | 0.042 | 0.015 | 0.073 | boundary | MID|IRF-3; | 0.0 |
| 9 | fp_D | Q14653 | 261 | VRHVLSCLGGGLALWR | HHHHHHCCCCCCEEEE | c1d-4 | multi | 0.036 | 0.015 | 0.069 | boundary | MID|IRF-3; | 0.0 |
| 10 | fp_D | Q14653 | 318 | GVFDLGPFIVDLITFTE | CCCCCHHHHHHHHHHHC | c1c-5 | multi-selected | 0.044 | 0.055 | 0.161 | boundary | MID|IRF-3; | 0.0 |
| 11 | fp_D | Q14653 | 318 | GVFDLGPFIVDLIT | CCCCCHHHHHHHHH | c3-4 | multi-selected | 0.024 | 0.053 | 0.161 | boundary | MID|IRF-3; | 0.0 |
| 12 | fp_D | Q14653 | 321 | DLGPFIVDLITFTE | CCHHHHHHHHHHHC | c2-5 | multi-selected | 0.051 | 0.053 | 0.141 | boundary | MID|IRF-3; | 0.0 |
| 13 | fp_D | Q14653 | 321 | DLGPFIVDLITFTE | CCHHHHHHHHHHHC | c3-4 | multi-selected | 0.051 | 0.053 | 0.141 | boundary | MID|IRF-3; | 0.0 |
| 14 | fp_beta_D | Q14653 | 355 | DQPWTKRLVMVKVVP | CCCCCCCEEEEEEEH | c1b-AT-4 | multi | 0.024 | 0.045 | 0.169 | boundary | boundary|IRF-3; | 0.571 |
| 15 | fp_beta_D | Q14653 | 360 | KRLVMVKVVPTCLRALV | CCEEEEEEEHHHHHHHH | c1cR-5 | multi | 0.014 | 0.022 | 0.138 | boundary | boundary|IRF-3; | 0.625 |
| 16 | fp_D | Q14653 | 364 | MVKVVPTCLRALVE | EEEEEHHHHHHHHH | c3-4 | multi-selected | 0.014 | 0.021 | 0.111 | boundary | boundary|IRF-3; | 0.286 |
| 17 | fp_D | Q14653 | 364 | MVKVVPTCLRALVEMAR | EEEEEHHHHHHHHHHHH | c1c-5 | multi-selected | 0.026 | 0.033 | 0.136 | boundary | boundary|IRF-3; | 0.125 |
| 18 | fp_D | Q14653 | 380 | RVGGASSLENTVDLHI | HHCCCCCCCCCCCCCC | c1d-AT-5 | multi-selected | 0.609 | 0.443 | 0.341 | boundary | boundary|IRF-3; | 0.0 |
| 19 | fp_D | Q14653 | 383 | GASSLENTVDLHISN | CCCCCCCCCCCCCCC | c1b-AT-4 | multi-selected | 0.725 | 0.496 | 0.37 | boundary | boundary|IRF-3; | 0.0 |
| 20 | fp_D | Q14653 | 408 | YKAYLQDLVEGMDFQG | HHHHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.398 | 0.385 | 0.281 | DISO | boundary|IRF-3; | 0.0 |
| 21 | fp_D | Q14653 | 408 | YKAYLQDLVEGMDFQG | HHHHHHHHHHHCCCCC | c1d-4 | multi | 0.398 | 0.385 | 0.281 | DISO | boundary|IRF-3; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment