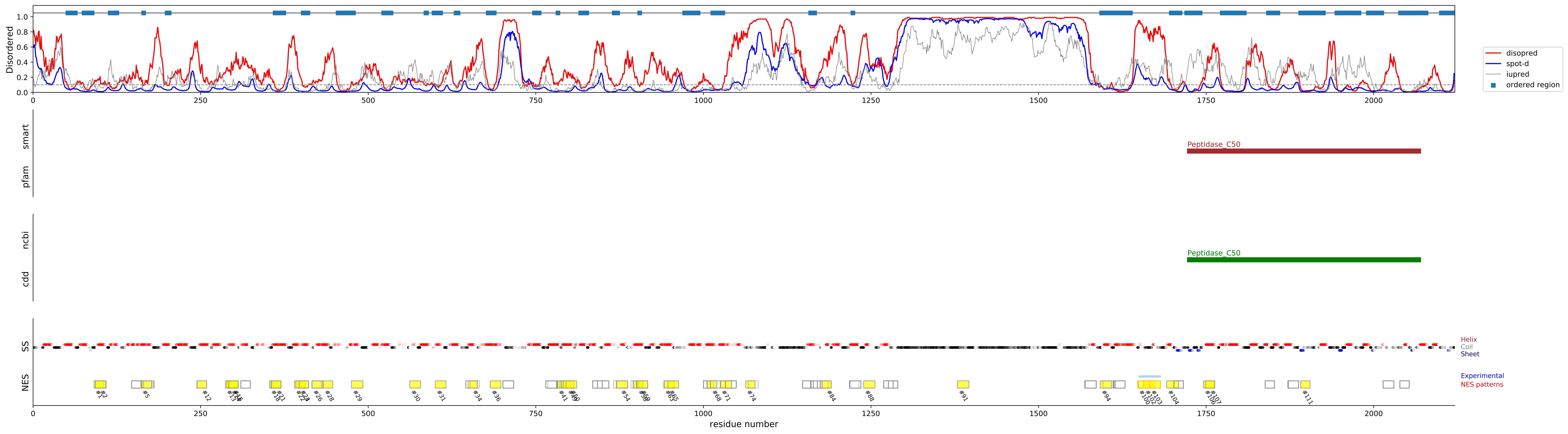

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14674 | 91 | QRPPLYLERILFVL | CCCCCHHHHHHHHH | c2-4 | multi-selected | 0.149 | 0.015 | 0.071 | boundary | 0.0 | |
| 2 | fp_D | Q14674 | 93 | PPLYLERILFVLLRNA | CCCHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.143 | 0.014 | 0.077 | boundary | 0.0 | |
| 3 | fp_D | Q14674 | 93 | PPLYLERILFVLLRN | CCCHHHHHHHHHHHH | c1b-4 | multi | 0.146 | 0.014 | 0.073 | boundary | 0.0 | |
| 4 | fp_D | Q14674 | 147 | SFSLLWKGAEALLE | HHHHHHHHHHHHHH | c3-AT | uniq | 0.225 | 0.032 | 0.059 | DISO | 0.0 | |
| 5 | fp_D | Q14674 | 161 | RRAAFAARLKALSFLV | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.183 | 0.027 | 0.038 | DISO | 0.0 | |
| 6 | fp_D | Q14674 | 162 | RAAFAARLKALSFLV | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.182 | 0.025 | 0.037 | DISO | 0.0 | |
| 7 | fp_D | Q14674 | 163 | AAFAARLKALSFLV | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.185 | 0.024 | 0.036 | DISO | 0.0 | |
| 8 | fp_D | Q14674 | 165 | FAARLKALSFLVLLED | HHHHHHHHHHHHHHCC | c1d-4 | multi | 0.245 | 0.026 | 0.041 | DISO | 0.0 | |
| 9 | fp_D | Q14674 | 165 | FAARLKALSFLVLLE | HHHHHHHHHHHHHHC | c1b-4 | multi | 0.229 | 0.024 | 0.038 | DISO | 0.0 | |
| 10 | fp_D | Q14674 | 165 | FAARLKALSFLVLL | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.214 | 0.022 | 0.035 | DISO | 0.0 | |
| 11 | fp_D | Q14674 | 244 | SPQRALCLLELTLEH | CCCHHHHHHHHHHHH | c1b-AT-4 | multi | 0.336 | 0.016 | 0.061 | DISO | 0.0 | |
| 12 | fp_D | Q14674 | 245 | PQRALCLLELTLEH | CCHHHHHHHHHHHH | c2-4 | multi-selected | 0.314 | 0.014 | 0.055 | DISO | 0.0 | |
| 13 | fp_D | Q14674 | 287 | LAPSLQLCQLGVKL | CCCHHHHHHHHHHH | c2-4 | multi-selected | 0.313 | 0.038 | 0.095 | DISO | 0.0 | |
| 14 | fp_D | Q14674 | 289 | PSLQLCQLGVKLLQVG | CHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.347 | 0.039 | 0.095 | DISO | 0.0 | |
| 15 | fp_D | Q14674 | 292 | QLCQLGVKLLQVGE | HHHHHHHHHHHHHC | c2-5 | multi-selected | 0.356 | 0.043 | 0.102 | DISO | 0.0 | |
| 16 | fp_D | Q14674 | 292 | QLCQLGVKLLQVGE | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.356 | 0.043 | 0.102 | DISO | 0.0 | |
| 17 | fp_D | Q14674 | 310 | AVAKLLIKASAVLS | HHHHHHHHHHHHHH | c3-AT | uniq | 0.381 | 0.088 | 0.12 | DISO | 0.0 | |
| 18 | fp_D | Q14674 | 353 | RRYRLDAILSLFAFLG | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.107 | 0.034 | 0.024 | __ | 0.0 | |
| 19 | fp_D | Q14674 | 353 | RRYRLDAILSLFAFLG | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.107 | 0.034 | 0.024 | __ | 0.0 | |
| 20 | fp_D | Q14674 | 355 | YRLDAILSLFAFLG | CCHHHHHHHHHHHH | c2-AT-4 | multi | 0.091 | 0.025 | 0.02 | __ | 0.0 | |
| 21 | fp_D | Q14674 | 356 | RLDAILSLFAFLGG | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.081 | 0.022 | 0.015 | boundary | 0.0 | |
| 22 | fp_D | Q14674 | 390 | QQSFLQMYFQGLHLYT | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.292 | 0.022 | 0.034 | boundary | 0.0 | |
| 23 | fp_D | Q14674 | 392 | SFLQMYFQGLHLYT | HHHHHHHHHHHHHH | c2-5 | multi-selected | 0.246 | 0.018 | 0.027 | boundary | 0.0 | |
| 24 | fp_D | Q14674 | 397 | YFQGLHLYTVVVYD | HHHHHHHHHHHHHH | c2-5 | multi-selected | 0.123 | 0.014 | 0.011 | boundary | 0.0 | |
| 25 | fp_D | Q14674 | 397 | YFQGLHLYTVVVYD | HHHHHHHHHHHHHH | c3-AT | multi | 0.123 | 0.014 | 0.011 | boundary | 0.0 | |
| 26 | fp_D | Q14674 | 416 | QIVDLADLTQLVDSCK | CCCCCHHHHHHHHHHH | c1aR-4 | multi-selected | 0.144 | 0.043 | 0.033 | boundary | 0.0 | |
| 27 | fp_D | Q14674 | 416 | QIVDLADLTQLVDS | CCCCCHHHHHHHHH | c3-AT | multi | 0.143 | 0.047 | 0.034 | boundary | 0.0 | |
| 28 | fp_D | Q14674 | 433 | TVVWMLEALEGLSG | HHHHHHHHHCCCCH | c3-4 | uniq | 0.431 | 0.038 | 0.075 | boundary | 0.0 | |
| 29 | fp_D | Q14674 | 475 | EACAISEPLCQHLGLVK | HHHHHHHHHHHHHHCCC | c1c-4 | uniq | 0.132 | 0.031 | 0.109 | boundary | 0.0 | |
| 30 | fp_D | Q14674 | 562 | KELQLKTLRDSLSGWD | CCHHHHHHHHHHCCCC | c1aR-4 | uniq | 0.154 | 0.068 | 0.303 | DISO | 0.0 | |
| 31 | fp_D | Q14674 | 600 | GQERFNIICDLLELSP | HHHHHHHHHHHHHCCC | c1d-4 | uniq | 0.134 | 0.033 | 0.178 | boundary | 0.0 | |
| 32 | fp_D | Q14674 | 645 | TNCSALDAIREALQLLD | CCCCHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.342 | 0.056 | 0.191 | boundary | 0.0 | |
| 33 | fp_D | Q14674 | 646 | NCSALDAIREALQLLD | CCCHHHHHHHHHHHHH | c1d-4 | multi | 0.343 | 0.051 | 0.196 | boundary | 0.0 | |
| 34 | fp_D | Q14674 | 649 | ALDAIREALQLLDS | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.384 | 0.042 | 0.212 | boundary | 0.0 | |
| 35 | fp_D | Q14674 | 652 | AIREALQLLDSVRP | HHHHHHHHHHHCCC | c3-AT | multi | 0.451 | 0.05 | 0.235 | __ | 0.0 | |
| 36 | fp_D | Q14674 | 682 | LWLYICTLEAKMQEGI | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.219 | 0.066 | 0.103 | boundary | 0.0 | |
| 37 | fp_D | Q14674 | 701 | RRAQAPGNLEEFEVND | HHCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.944 | 0.722 | 0.42 | boundary | 0.0 | |
| 38 | fp_D | Q14674 | 765 | AVRCLQQTAASLQILA | CCCCHHHHHHHHHHHH | c1a-AT-5 | multi-selected | 0.285 | 0.025 | 0.079 | boundary | 0.0 | |
| 39 | fp_D | Q14674 | 765 | AVRCLQQTAASLQILA | CCCCHHHHHHHHHHHH | c1d-AT-5 | multi | 0.285 | 0.025 | 0.079 | boundary | 0.0 | |
| 40 | fp_D | Q14674 | 768 | CLQQTAASLQILAA | CHHHHHHHHHHHHH | c3-AT | multi | 0.251 | 0.019 | 0.056 | boundary | 0.0 | |
| 41 | fp_D | Q14674 | 782 | LYQLVAKPMQALEVLL | HHHHHCCHHHHHHHHH | c1a-4 | multi-selected | 0.187 | 0.016 | 0.024 | DISO | 0.0 | |
| 42 | fp_D | Q14674 | 783 | YQLVAKPMQALEVLL | HHHHCCHHHHHHHHH | c1b-AT-4 | multi | 0.191 | 0.017 | 0.023 | DISO | 0.0 | |
| 43 | fp_D | Q14674 | 786 | VAKPMQALEVLLLL | HCCHHHHHHHHHHH | c2-AT-4 | multi | 0.217 | 0.016 | 0.028 | DISO | 0.0 | |
| 44 | fp_D | Q14674 | 786 | VAKPMQALEVLLLLRI | HCCHHHHHHHHHHHHH | c1d-4 | multi | 0.218 | 0.016 | 0.031 | boundary | 0.0 | |
| 45 | fp_D | Q14674 | 786 | VAKPMQALEVLLLLR | HCCHHHHHHHHHHHH | c1b-4 | multi | 0.217 | 0.016 | 0.029 | boundary | 0.0 | |
| 46 | fp_D | Q14674 | 788 | KPMQALEVLLLLRIVS | CHHHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.226 | 0.017 | 0.034 | boundary | 0.0 | |
| 47 | fp_D | Q14674 | 788 | KPMQALEVLLLLRIVS | CHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.226 | 0.017 | 0.034 | boundary | 0.0 | |
| 48 | fp_D | Q14674 | 789 | PMQALEVLLLLRIVS | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.229 | 0.017 | 0.036 | boundary | 0.0 | |
| 49 | fp_D | Q14674 | 791 | QALEVLLLLRIVSERL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.234 | 0.021 | 0.046 | boundary | 0.0 | |
| 50 | fp_D | Q14674 | 795 | VLLLLRIVSERLKDHS | HHHHHHHHHHHHCCHH | c1aR-4 | multi-selected | 0.21 | 0.036 | 0.076 | boundary | 0.0 | |
| 51 | fp_D | Q14674 | 835 | HLEEAASSLKHLDQ | HHHHHHHHCCCCCC | c3-AT | uniq | 0.531 | 0.133 | 0.166 | boundary | 0.0 | |
| 52 | fp_D | Q14674 | 842 | SLKHLDQTTDTYLLLSL | HCCCCCCCCCHHHHHHH | c1c-AT-5 | uniq | 0.509 | 0.119 | 0.097 | boundary | 0.0 | |
| 53 | fp_D | Q14674 | 866 | QLYWTHQKVTKGVSLLL | HHHHHCCCHHHHHHHHH | c1c-AT-5 | multi | 0.14 | 0.024 | 0.045 | DISO | 0.0 | |
| 54 | fp_D | Q14674 | 870 | THQKVTKGVSLLLSVLR | HCCCHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.215 | 0.028 | 0.055 | boundary | 0.0 | |
| 55 | fp_D | Q14674 | 871 | HQKVTKGVSLLLSVLR | CCCHHHHHHHHHHHHC | c1d-AT-4 | multi | 0.222 | 0.029 | 0.056 | boundary | 0.0 | |
| 56 | fp_D | Q14674 | 892 | KSSKAWYLLRVQVLQ | CCCHHHHHHHHHHHH | c1b-AT-4 | multi | 0.228 | 0.021 | 0.022 | DISO | 0.0 | |
| 57 | fp_D | Q14674 | 895 | KAWYLLRVQVLQLVA | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.17 | 0.012 | 0.017 | DISO | 0.0 | |
| 58 | fp_D | Q14674 | 896 | AWYLLRVQVLQLVA | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.156 | 0.011 | 0.016 | DISO | 0.0 | |
| 59 | fp_D | Q14674 | 900 | LRVQVLQLVAAYLSLPS | HHHHHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.165 | 0.016 | 0.02 | DISO | 0.0 | |
| 60 | fp_D | Q14674 | 901 | RVQVLQLVAAYLSLPS | HHHHHHHHHHHHCCCC | c1a-AT-5 | multi | 0.163 | 0.016 | 0.021 | DISO | 0.0 | |
| 61 | fp_D | Q14674 | 901 | RVQVLQLVAAYLSLPS | HHHHHHHHHHHHCCCC | c1d-5 | multi | 0.163 | 0.016 | 0.021 | DISO | 0.0 | |
| 62 | fp_D | Q14674 | 903 | QVLQLVAAYLSLPS | HHHHHHHHHHCCCC | c2-AT-5 | multi | 0.163 | 0.017 | 0.022 | DISO | 0.0 | |
| 63 | fp_D | Q14674 | 941 | DSHKLLRSIILLLMGS | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.205 | 0.019 | 0.013 | __ | 0.0 | |
| 64 | fp_D | Q14674 | 942 | SHKLLRSIILLLMGS | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.203 | 0.019 | 0.012 | __ | 0.0 | |
| 65 | fp_D | Q14674 | 947 | RSIILLLMGSDILSTQ | HHHHHHHHCCCCCCCC | c1aR-4 | multi-selected | 0.222 | 0.068 | 0.018 | boundary | 0.0 | |
| 66 | fp_D | Q14674 | 1000 | RLGSVSEAKAFCLEA | HCCCHHHHHHHHHHH | c1b-AT-5 | multi-selected | 0.145 | 0.033 | 0.04 | boundary | 0.0 | |
| 67 | fp_D | Q14674 | 1005 | SEAKAFCLEALKLTT | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.097 | 0.03 | 0.047 | boundary | 0.0 | |
| 68 | fp_D | Q14674 | 1006 | EAKAFCLEALKLTT | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.093 | 0.03 | 0.049 | boundary | 0.0 | |
| 69 | fp_D | Q14674 | 1010 | FCLEALKLTTKLQIPR | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.06 | 0.031 | 0.04 | boundary | 0.0 | |
| 70 | fp_D | Q14674 | 1011 | CLEALKLTTKLQIPR | HHHHHHHHHHHHHHH | c1b-AT-5 | multi | 0.055 | 0.031 | 0.038 | boundary | 0.0 | |
| 71 | fp_D | Q14674 | 1025 | RQCALFLVLKGELELAR | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.244 | 0.036 | 0.041 | boundary | 0.0 | |
| 72 | fp_D | Q14674 | 1026 | QCALFLVLKGELELAR | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.257 | 0.035 | 0.042 | boundary | 0.0 | |
| 73 | fp_D | Q14674 | 1033 | LKGELELARNDIDLCQ | HHHHHHHHHHHHHHHH | c1d-AT-4 | uniq | 0.494 | 0.074 | 0.077 | boundary | 0.0 | |
| 74 | fp_D | Q14674 | 1063 | EFGGVTQHLDSVKK | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.866 | 0.491 | 0.189 | DISO | 0.0 | |
| 75 | fp_D | Q14674 | 1067 | VTQHLDSVKKVHLQK | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.921 | 0.565 | 0.264 | DISO | 0.0 | |
| 76 | fp_D | Q14674 | 1148 | CSLCASPVLTAVCLRW | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.16 | 0.124 | 0.04 | boundary | 0.0 | |
| 77 | fp_D | Q14674 | 1148 | CSLCASPVLTAVCLRW | CCCCCCHHHHHHHHHH | c1d-AT-4 | multi | 0.16 | 0.124 | 0.04 | boundary | 0.0 | |
| 78 | fp_D | Q14674 | 1160 | CLRWVLVTAGVRLAMGH | HHHHHHHHHHHHHCCCC | c1c-AT-5 | multi-selected | 0.144 | 0.122 | 0.084 | boundary | 0.0 | |
| 79 | fp_D | Q14674 | 1160 | CLRWVLVTAGVRLAM | HHHHHHHHHHHHHCC | c1b-AT-5 | multi | 0.125 | 0.114 | 0.07 | boundary | 0.0 | |
| 80 | fp_D | Q14674 | 1161 | LRWVLVTAGVRLAMGH | HHHHHHHHHHHHCCCC | c1d-AT-4 | multi | 0.149 | 0.124 | 0.088 | boundary | 0.0 | |
| 81 | fp_D | Q14674 | 1161 | LRWVLVTAGVRLAM | HHHHHHHHHHHHCC | c2-AT-4 | multi | 0.13 | 0.115 | 0.074 | boundary | 0.0 | |
| 82 | fp_D | Q14674 | 1170 | VRLAMGHQAQGLDLLQ | HHHCCCCCHHHHHHHH | c1a-AT-4 | multi-selected | 0.324 | 0.144 | 0.153 | boundary | 0.0 | |
| 83 | fp_D | Q14674 | 1174 | MGHQAQGLDLLQVVL | CCCCHHHHHHHHHHH | c1b-AT-4 | multi | 0.384 | 0.126 | 0.165 | boundary | 0.0 | |
| 84 | fp_D | Q14674 | 1177 | QAQGLDLLQVVLKG | CHHHHHHHHHHHHC | c2-4 | multi-selected | 0.434 | 0.105 | 0.164 | boundary | 0.0 | |
| 85 | fp_D | Q14674 | 1218 | LLDEILAQAYTLLALEG | HHHHHHHHHHHHHHHHC | c1c-AT-5 | multi-selected | 0.224 | 0.084 | 0.129 | DISO | 0.0 | |
| 86 | fp_D | Q14674 | 1218 | LLDEILAQAYTLLA | HHHHHHHHHHHHHH | c3-AT | multi | 0.174 | 0.081 | 0.112 | DISO | 0.0 | |
| 87 | fp_D | Q14674 | 1219 | LDEILAQAYTLLALEG | HHHHHHHHHHHHHHHC | c1d-AT-4 | multi | 0.224 | 0.083 | 0.121 | DISO | 0.0 | |
| 88 | fp_D | Q14674 | 1239 | SNESLQKVLQSGLKFVA | CHHHHHHHHHHCCCHHC | c1c-4 | uniq | 0.524 | 0.322 | 0.245 | DISO | 0.0 | |
| 89 | fp_D | Q14674 | 1269 | LLIWALTKLGGLSC | HHHHHHHHHCCCCC | c3-AT | uniq | 0.275 | 0.174 | 0.057 | DISO | 0.0 | |
| 90 | fp_D | Q14674 | 1276 | KLGGLSCCTTQLFA | HHCCCCCCCCCCCC | c3-AT | uniq | 0.427 | 0.343 | 0.1 | DISO | 0.0 | |
| 91 | fp_D | Q14674 | 1379 | QAPRVQQRVQTRLKVNF | CCCCCCCCCCCCCCCCC | c1c-4 | uniq | 0.982 | 0.948 | 0.753 | DISO | 0.0 | |
| 92 | fp_D | Q14674 | 1569 | TGLSTLDSICDSLSVAF | CCCCCHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.503 | 0.212 | 0.222 | boundary | 0.0 | |
| 93 | fp_D | Q14674 | 1570 | GLSTLDSICDSLSVAF | CCCCHHHHHHHHHHHH | c1d-5 | multi | 0.477 | 0.189 | 0.215 | boundary | 0.0 | |
| 94 | fp_D | Q14674 | 1592 | PPSGLYAHLCRFLALCL | CCCCCHHHHHHHHHHHH | c1c-4 | multi-selected | 0.064 | 0.06 | 0.062 | boundary | 0.0 | |
| 95 | fp_D | Q14674 | 1596 | LYAHLCRFLALCLGH | CHHHHHHHHHHHHCC | c1b-4 | multi | 0.067 | 0.052 | 0.051 | boundary | 0.0 | |
| 96 | fp_D | Q14674 | 1612 | DPYATAFLVTESVSITC | CHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.02 | 0.037 | 0.117 | boundary | 0.0 | |
| 97 | fp_D | Q14674 | 1613 | PYATAFLVTESVSITC | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.018 | 0.036 | 0.119 | boundary | 0.0 | |
| 98 | fp_D | Q14674 | 1614 | YATAFLVTESVSITC | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.016 | 0.036 | 0.123 | boundary | 0.0 | |
| 99 | fp_D | Q14674 | 1615 | ATAFLVTESVSITC | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.015 | 0.036 | 0.125 | boundary | 0.0 | |
| 100 | fp_D | Q14674 | 1648 | GSLEIADQLQGLSLQE ............. | CCHHHHHHHCCCCCCC | c1a-4 | multi-selected | 0.869 | 0.24 | 0.347 | boundary | 0.0 | |
| 101 | fp_D | Q14674 | 1649 | SLEIADQLQGLSLQE ............. | CHHHHHHHCCCCCCC | c1b-AT-5 | multi | 0.867 | 0.23 | 0.342 | boundary | 0.0 | |
| 102 | fp_D | Q14674 | 1657 | QGLSLQEMPGDVPLAR ................ | CCCCCCCCCCCCCHHH | c1aR-4 | uniq | 0.781 | 0.155 | 0.273 | DISO | 0.0 | |
| 103 | fp_D | Q14674 | 1666 | GDVPLARIQRLFSFRA ................ | CCCCHHHHHHHCCCCC | c1d-4 | uniq | 0.79 | 0.116 | 0.176 | DISO | 0.0 | |
| 104 | fp_D | Q14674 | 1691 | EKESFQERLALIPSGVTV | HHHHHHHHHHCCCCCEEE | c4-4 | uniq | 0.106 | 0.039 | 0.226 | boundary | boundary|Peptidase_C50; | 0.0 |
| 105 | fp_beta_D | Q14674 | 1702 | IPSGVTVCVLALAT | CCCCEEEEEEEECC | c2-4 | uniq | 0.067 | 0.016 | 0.105 | boundary | boundary|Peptidase_C50; | 0.857 |
| 106 | fp_D | Q14674 | 1746 | NKLHLRSVLNEFDAIQ | CCCHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.387 | 0.079 | 0.299 | boundary | MID|Peptidase_C50; | 0.0 |
| 107 | fp_D | Q14674 | 1749 | HLRSVLNEFDAIQK | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.45 | 0.071 | 0.303 | boundary | MID|Peptidase_C50; | 0.0 |
| 108 | fp_D | Q14674 | 1838 | LLKIMLSGAGALTP | HHHHHHHHCCCCCH | c3-AT | uniq | 0.104 | 0.031 | 0.184 | __ | MID|Peptidase_C50; | 0.0 |
| 109 | fp_D | Q14674 | 1872 | LLNEAVGRLQGLTVPS | HHHHHHHHHCCCCCCC | c1a-AT-5 | multi-selected | 0.319 | 0.081 | 0.323 | boundary | MID|Peptidase_C50; | 0.0 |
| 110 | fp_D | Q14674 | 1873 | LNEAVGRLQGLTVPS | HHHHHHHHCCCCCCC | c1b-4 | multi | 0.315 | 0.084 | 0.313 | boundary | MID|Peptidase_C50; | 0.0 |
| 111 | fp_D | Q14674 | 1891 | LVLVLDKDLQKLPW | EEEEECCCCCCCCC | c3-4 | uniq | 0.019 | 0.017 | 0.22 | boundary | MID|Peptidase_C50; | 0.286 |
| 112 | fp_D | Q14674 | 2014 | AVLRLSCRAVALLFGC | HHHCCCCCCCEEEECC | c1a-AT-5 | uniq | 0.296 | 0.022 | 0.019 | boundary | MID|Peptidase_C50; | 0.143 |
| 113 | fp_D | Q14674 | 2039 | NLEGAGIVLKYIMA | CCCCCCHHHHHHHH | c3-AT | uniq | 0.026 | 0.018 | 0.024 | boundary | boundary|Peptidase_C50; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment