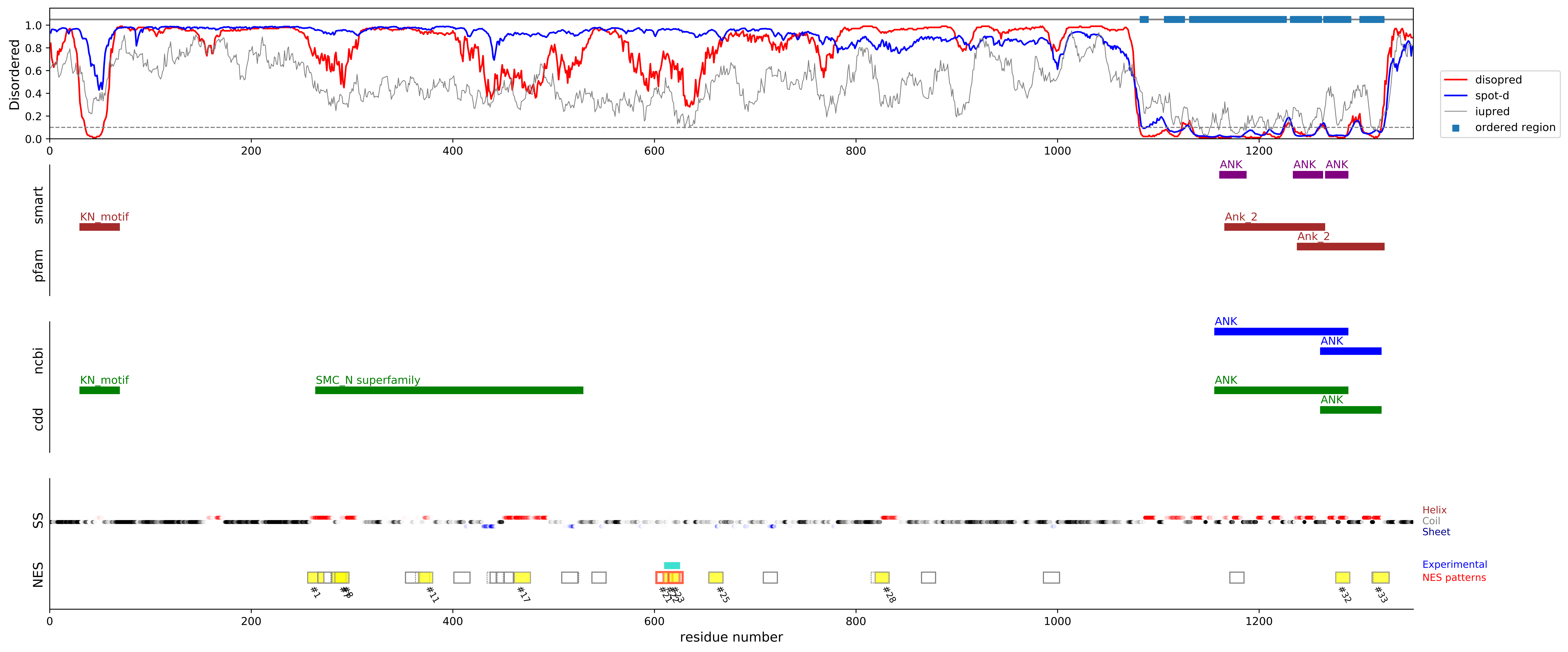

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14678 | 256 | SPMHLQHIREQMAIAL | CCCHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.755 | 0.963 | 0.49 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 2 | fp_D | Q14678 | 256 | SPMHLQHIREQMAIAL | CCCHHHHHHHHHHHHH | c1d-4 | multi | 0.755 | 0.963 | 0.49 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 3 | fp_D | Q14678 | 266 | QMAIALKRLKELEE | HHHHHHHHHHHHHH | c3-AT | uniq | 0.689 | 0.939 | 0.396 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 4 | fp_D | Q14678 | 279 | EQVRTIPVLQVKISVLQ | HHHCCCCCCHHHHHHHH | c1c-AT-4 | multi | 0.614 | 0.944 | 0.326 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 5 | fp_D | Q14678 | 279 | EQVRTIPVLQVKISV | HHHCCCCCCHHHHHH | c1b-AT-4 | multi | 0.609 | 0.944 | 0.329 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 6 | fp_D | Q14678 | 280 | QVRTIPVLQVKISVLQ | HHCCCCCCHHHHHHHH | c1d-5 | multi | 0.616 | 0.946 | 0.329 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 7 | fp_D | Q14678 | 280 | QVRTIPVLQVKISV | HHCCCCCCHHHHHH | c2-5 | multi-selected | 0.611 | 0.945 | 0.332 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 8 | fp_D | Q14678 | 283 | TIPVLQVKISVLQE | CCCCCHHHHHHHHH | c3-4 | multi-selected | 0.619 | 0.95 | 0.321 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 9 | fp_D | Q14678 | 353 | EMETVEQSTQRIKE | HHHHHCCCCCCHHH | c3-AT | uniq | 0.966 | 0.977 | 0.49 | DISO | MID|SMC_N superfamily; | 0.0 |
| 10 | fp_D | Q14678 | 363 | RIKEFRQLTADMQA | CHHHHCCCHHHHHH | c3-AT | multi | 0.941 | 0.969 | 0.415 | DISO | MID|SMC_N superfamily; | 0.0 |
| 11 | fp_D | Q14678 | 366 | EFRQLTADMQALEQ | HHCCCHHHHHHHHH | c3-4 | multi-selected | 0.938 | 0.966 | 0.398 | DISO | MID|SMC_N superfamily; | 0.0 |
| 12 | fp_D | Q14678 | 401 | VAVGAEENMNDIVVYH | CCCCCCCCCCCEEEEE | c1a-AT-4 | uniq | 0.837 | 0.959 | 0.426 | DISO | MID|SMC_N superfamily; | 0.0 |
| 13 | fp_beta_D | Q14678 | 434 | RNCGVSVTEAMLGVMT | EEEEEEEEEECCCCCC | c1d-AT-4 | multi | 0.469 | 0.837 | 0.371 | DISO | MID|SMC_N superfamily; | 0.857 |
| 14 | fp_beta_D | Q14678 | 437 | GVSVTEAMLGVMTE | EEEEEEECCCCCCH | c3-AT | multi-selected | 0.484 | 0.821 | 0.386 | DISO | MID|SMC_N superfamily; | 0.571 |
| 15 | fp_D | Q14678 | 443 | AMLGVMTEADKEIELQQ | ECCCCCCHHHHHHHHHH | c1c-AT-5 | multi-selected | 0.541 | 0.883 | 0.443 | DISO | MID|SMC_N superfamily; | 0.0 |
| 16 | fp_D | Q14678 | 444 | MLGVMTEADKEIELQQ | CCCCCCHHHHHHHHHH | c1d-AT-5 | multi | 0.541 | 0.888 | 0.445 | DISO | MID|SMC_N superfamily; | 0.0 |
| 17 | fp_D | Q14678 | 461 | TIESLKEKIYRLEVQL | HHHHHHHHHHHHHHHH | c1a-5 | uniq | 0.504 | 0.923 | 0.345 | DISO | MID|SMC_N superfamily; | 0.0 |
| 18 | fp_beta_D | Q14678 | 508 | QPLVFSKVVEAVVQTRD | CCCEEEEEEEEEEEEEC | c1cR-4 | multi | 0.639 | 0.931 | 0.308 | DISO | boundary|SMC_N superfamily; | 1.0 |
| 19 | fp_beta_D | Q14678 | 508 | QPLVFSKVVEAVVQTR | CCCEEEEEEEEEEEEE | c1aR-4 | multi-selected | 0.636 | 0.932 | 0.305 | DISO | boundary|SMC_N superfamily; | 1.0 |
| 20 | fp_D | Q14678 | 538 | VGTSVETNSVGISC | CCCCCCCCCEEECC | c2-AT-4 | uniq | 0.959 | 0.97 | 0.315 | DISO | boundary|SMC_N superfamily; | 0.143 |
| 21 | cand_D | Q14678 | 602 | TGSNTEESVNDLTLLK +++++ | CCCCCCEEECCCCHHC | c1a-AT-4 | uniq | 0.652 | 0.958 | 0.425 | DISO | 0.429 | |
| 22 | cand_D | Q14678 | 609 | SVNDLTLLKTNLNLKE ++++++++++ | EECCCCHHCCCCCCCC | c1d-5 | multi-selected | 0.666 | 0.963 | 0.351 | DISO | 0.0 | |
| 23 | cand_D | Q14678 | 614 | TLLKTNLNLKEVRS +++++++++ | CHHCCCCCCCCEEE | c3-AT | multi-selected | 0.622 | 0.961 | 0.279 | DISO | 0.0 | |
| 24 | fp_D | Q14678 | 654 | AVSQVEAAVMAVPR | CCCCCCCEEEECCC | c2-AT-5 | multi | 0.776 | 0.928 | 0.518 | DISO | 0.429 | |
| 25 | fp_D | Q14678 | 654 | AVSQVEAAVMAVPR | CCCCCCCEEEECCC | c3-4 | multi-selected | 0.776 | 0.928 | 0.518 | DISO | 0.429 | |
| 26 | fp_beta_D | Q14678 | 708 | STQTVETRTVAVGE | CCCCEEEEEEEECC | c2-AT-4 | uniq | 0.86 | 0.949 | 0.562 | DISO | 0.857 | |
| 27 | fp_D | Q14678 | 815 | APLGMMTGLDHYIERIQ | CCCCCCCCCCHHHHHHH | c1cR-5 | multi | 0.957 | 0.824 | 0.454 | DISO | 0.0 | |
| 28 | fp_D | Q14678 | 819 | MMTGLDHYIERIQK | CCCCCCHHHHHHHH | c3-4 | multi-selected | 0.948 | 0.819 | 0.395 | DISO | 0.0 | |
| 29 | fp_D | Q14678 | 865 | QLISTLSSINSVMK | CCCCCCCCCCCCCC | c3-AT | uniq | 0.977 | 0.887 | 0.415 | DISO | 0.0 | |
| 30 | fp_D | Q14678 | 986 | DSNGAKKNLQFVGING | CCCCCCCCEEEECCCC | c1a-AT-4 | uniq | 0.859 | 0.779 | 0.53 | DISO | 0.429 | |
| 31 | fp_O | Q14678 | 1171 | SHSNFEIVKLLLDA | HCCCHHHHHHHHCC | c2-4 | uniq | 0.019 | 0.02 | 0.072 | ORD | boundary|ANK; | 0.0 |
| 32 | fp_D | Q14678 | 1276 | EHGHVEIVKLLLAQ | HCCCHHHHHHHHCC | c2-4 | uniq | 0.026 | 0.03 | 0.208 | boundary | boundary|ANK; boundary|ANK; | 0.0 |
| 33 | fp_D | Q14678 | 1312 | GHKDIAVLLYAHVNFAK | CCHHHHHHHHHCCCCCC | c1c-4 | multi-selected | 0.195 | 0.122 | 0.158 | boundary | boundary|ANK; boundary|ANK; | 0.0 |
| 34 | fp_D | Q14678 | 1313 | HKDIAVLLYAHVNFAK | CHHHHHHHHHCCCCCC | c1d-AT-4 | multi | 0.206 | 0.126 | 0.162 | boundary | boundary|ANK; boundary|ANK; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment