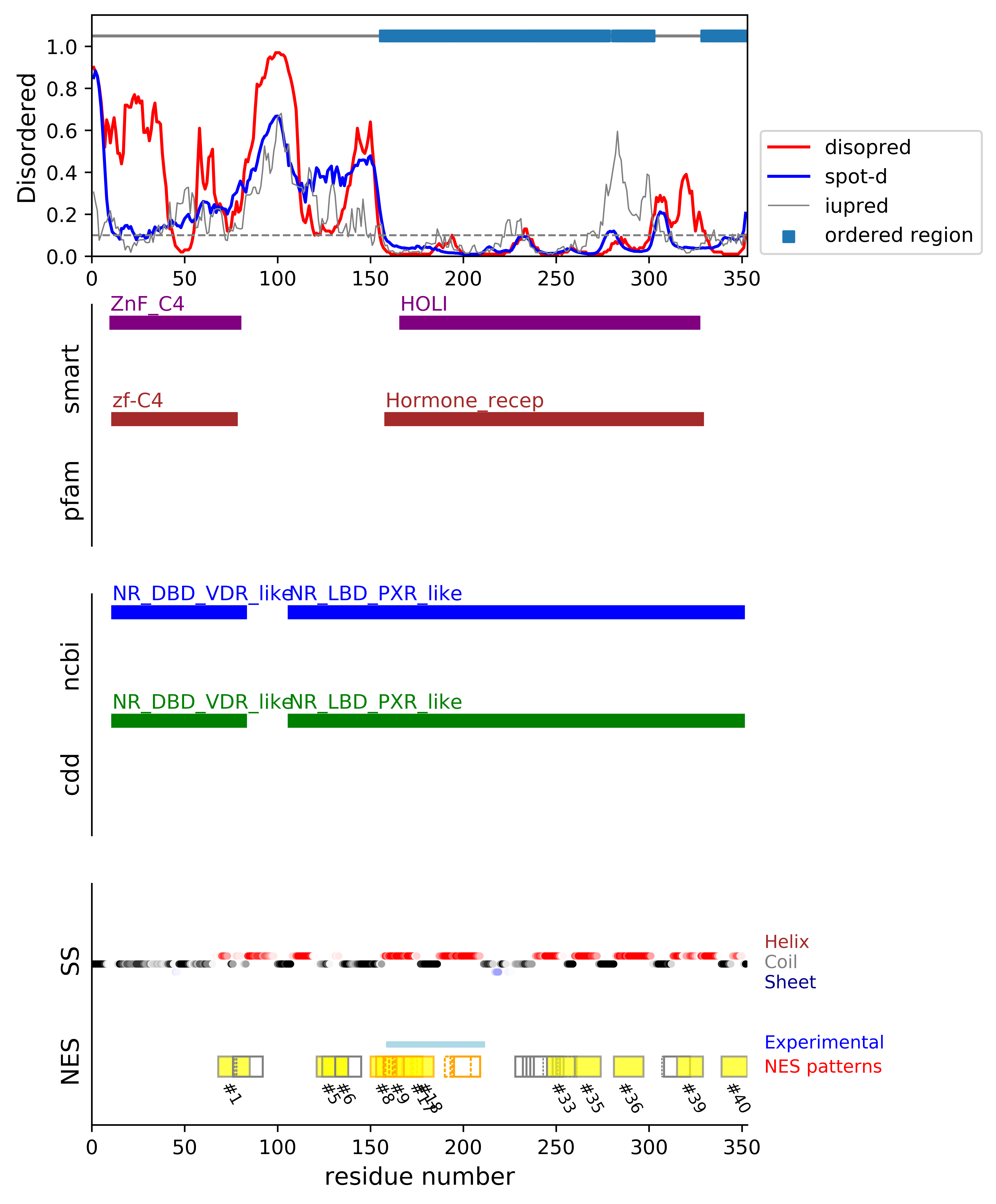

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q14994 | 68 | LQKCLDAGMRKDMILSA | HHHHHHCCCHHHHCCCH | c1c-4 | uniq | 0.246 | 0.277 | 0.165 | DISO | boundary|NR_DBD_VDR_like; | 0.0 |
| 2 | fp_D | Q14994 | 76 | MRKDMILSAEALALRR | CHHHHCCCHHHHHHHH | c1a-AT-4 | multi-selected | 0.463 | 0.372 | 0.237 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD_PXR_like; | 0.0 |
| 3 | fp_D | Q14994 | 77 | RKDMILSAEALALRR | HHHHCCCHHHHHHHH | c1b-AT-4 | multi | 0.48 | 0.378 | 0.244 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD_PXR_like; | 0.0 |
| 4 | fp_D | Q14994 | 78 | KDMILSAEALALRR | HHHCCCHHHHHHHH | c2-AT-4 | multi | 0.5 | 0.384 | 0.25 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD_PXR_like; | 0.0 |
| 5 | fp_D | Q14994 | 121 | HTRHMGTMFEQFVQFRP | CCCCCCCCHHHHHCCCC | c1c-4 | multi-selected | 0.149 | 0.386 | 0.195 | DISO | boundary|NR_LBD_PXR_like; | 0.0 |
| 6 | fp_D | Q14994 | 124 | HMGTMFEQFVQFRP | CCCCCHHHHHCCCC | c3-4 | multi-selected | 0.159 | 0.383 | 0.196 | DISO | boundary|NR_LBD_PXR_like; | 0.0 |
| 7 | fp_D | Q14994 | 131 | QFVQFRPPAHLFIH | HHHCCCCCCCCCCC | c3-AT | uniq | 0.327 | 0.399 | 0.161 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 8 | fp_D | Q14994 | 150 | TLAPVLPLVTHFAD .... | CCCCCCCHHHHHHH | c3-4 | multi-selected | 0.166 | 0.209 | 0.077 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 9 | fp_D | Q14994 | 153 | PVLPLVTHFADINT ....... | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.056 | 0.122 | 0.064 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 10 | fp_D | Q14994 | 157 | LVTHFADINTFMVLQ ............ | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.015 | 0.056 | 0.029 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 11 | fp_D | Q14994 | 157 | LVTHFADINTFMVLQV ............. | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.014 | 0.055 | 0.028 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 12 | fp_D | Q14994 | 158 | VTHFADINTFMVLQ ............ | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.013 | 0.051 | 0.026 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 13 | fp_D | Q14994 | 160 | HFADINTFMVLQVIK ............... | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.011 | 0.045 | 0.021 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 14 | fp_D | Q14994 | 162 | ADINTFMVLQVIKFTK ................ | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi | 0.01 | 0.042 | 0.02 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 15 | fp_D | Q14994 | 162 | ADINTFMVLQVIKFTK ................ | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.01 | 0.042 | 0.02 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 16 | fp_D | Q14994 | 163 | DINTFMVLQVIKFTK ............... | HHHHHHHHHHHHHCC | c1b-5 | multi | 0.01 | 0.042 | 0.02 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 17 | fp_D | Q14994 | 164 | INTFMVLQVIKFTK .............. | HHHHHHHHHHHHCC | c2-4 | multi-selected | 0.01 | 0.041 | 0.02 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 18 | fp_D | Q14994 | 168 | MVLQVIKFTKDLPVFR ................ | HHHHHHHHCCCCCCCC | c1aR-5 | multi-selected | 0.011 | 0.04 | 0.033 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 19 | fp_O | Q14994 | 190 | QISLLKGAAVEICH .............. | HHHHHHHHHHHHHH | c3-AT | multi | 0.045 | 0.014 | 0.042 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 20 | fp_O | Q14994 | 193 | LLKGAAVEICHIVLNT ................ | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.031 | 0.012 | 0.035 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 21 | fp_O | Q14994 | 194 | LKGAAVEICHIVLNT ............... | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.028 | 0.011 | 0.031 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 22 | fp_O | Q14994 | 195 | KGAAVEICHIVLNT .............. | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.023 | 0.011 | 0.03 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 23 | fp_O | Q14994 | 228 | DGARVSPTVGFQVEFLE | CCCCCCCCCCHHHHHHH | c1c-4 | multi-selected | 0.063 | 0.058 | 0.069 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 24 | fp_O | Q14994 | 228 | DGARVSPTVGFQVEF | CCCCCCCCCCHHHHH | c1b-AT-4 | multi | 0.07 | 0.064 | 0.076 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 25 | fp_O | Q14994 | 232 | VSPTVGFQVEFLELLF | CCCCCCHHHHHHHHHH | c1a-4 | multi-selected | 0.051 | 0.044 | 0.038 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 26 | fp_O | Q14994 | 234 | PTVGFQVEFLELLFHFH | CCCCHHHHHHHHHHHHH | c1cR-5 | multi | 0.036 | 0.033 | 0.029 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 27 | fp_O | Q14994 | 234 | PTVGFQVEFLELLFHF | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.038 | 0.034 | 0.03 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 28 | fp_O | Q14994 | 234 | PTVGFQVEFLELLF | CCCCHHHHHHHHHH | c2-4 | multi-selected | 0.042 | 0.037 | 0.032 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 29 | fp_O | Q14994 | 236 | VGFQVEFLELLFHFHG | CCHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.026 | 0.025 | 0.023 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 30 | fp_O | Q14994 | 236 | VGFQVEFLELLFHFHG | CCHHHHHHHHHHHHHH | c1d-4 | multi | 0.026 | 0.025 | 0.023 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 31 | fp_O | Q14994 | 236 | VGFQVEFLELLFHF | CCHHHHHHHHHHHH | c2-4 | multi-selected | 0.028 | 0.026 | 0.022 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 32 | fp_O | Q14994 | 238 | FQVEFLELLFHFHGTL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.018 | 0.02 | 0.022 | ORD | MID|NR_LBD_PXR_like; | 0.0 |
| 33 | fp_D | Q14994 | 245 | LLFHFHGTLRKLQLQE | HHHHHHHHHHCCCCCH | c1a-5 | multi-selected | 0.019 | 0.022 | 0.031 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 34 | fp_D | Q14994 | 245 | LLFHFHGTLRKLQLQE | HHHHHHHHHHCCCCCH | c1d-AT-5 | multi | 0.019 | 0.022 | 0.031 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 35 | fp_D | Q14994 | 260 | EPEYVLLAAMALFS | HHHHHHHHHHHHHC | c2-4 | uniq | 0.016 | 0.016 | 0.074 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 36 | fp_D | Q14994 | 281 | QRDEIDQLQEEMALTL | CHHHHHHHHHHHHHHH | c1d-4 | uniq | 0.053 | 0.048 | 0.321 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 37 | fp_D | Q14994 | 307 | PRDRFLYAKLLGLLA | CCCCCCHHHHHHHHH | c1b-AT-4 | multi | 0.251 | 0.088 | 0.064 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 38 | fp_D | Q14994 | 308 | RDRFLYAKLLGLLA | CCCCCHHHHHHHHH | c2-AT-4 | multi-selected | 0.251 | 0.079 | 0.061 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 39 | fp_D | Q14994 | 315 | KLLGLLAELRSINE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.26 | 0.04 | 0.034 | boundary | MID|NR_LBD_PXR_like; | 0.0 |
| 40 | fp_D | Q14994 | 339 | GLSAMMPLLQEICS | CCCCCCHHHHHHCC | c3-4 | uniq | 0.021 | 0.096 | 0.074 | boundary | boundary|NR_LBD_PXR_like; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment