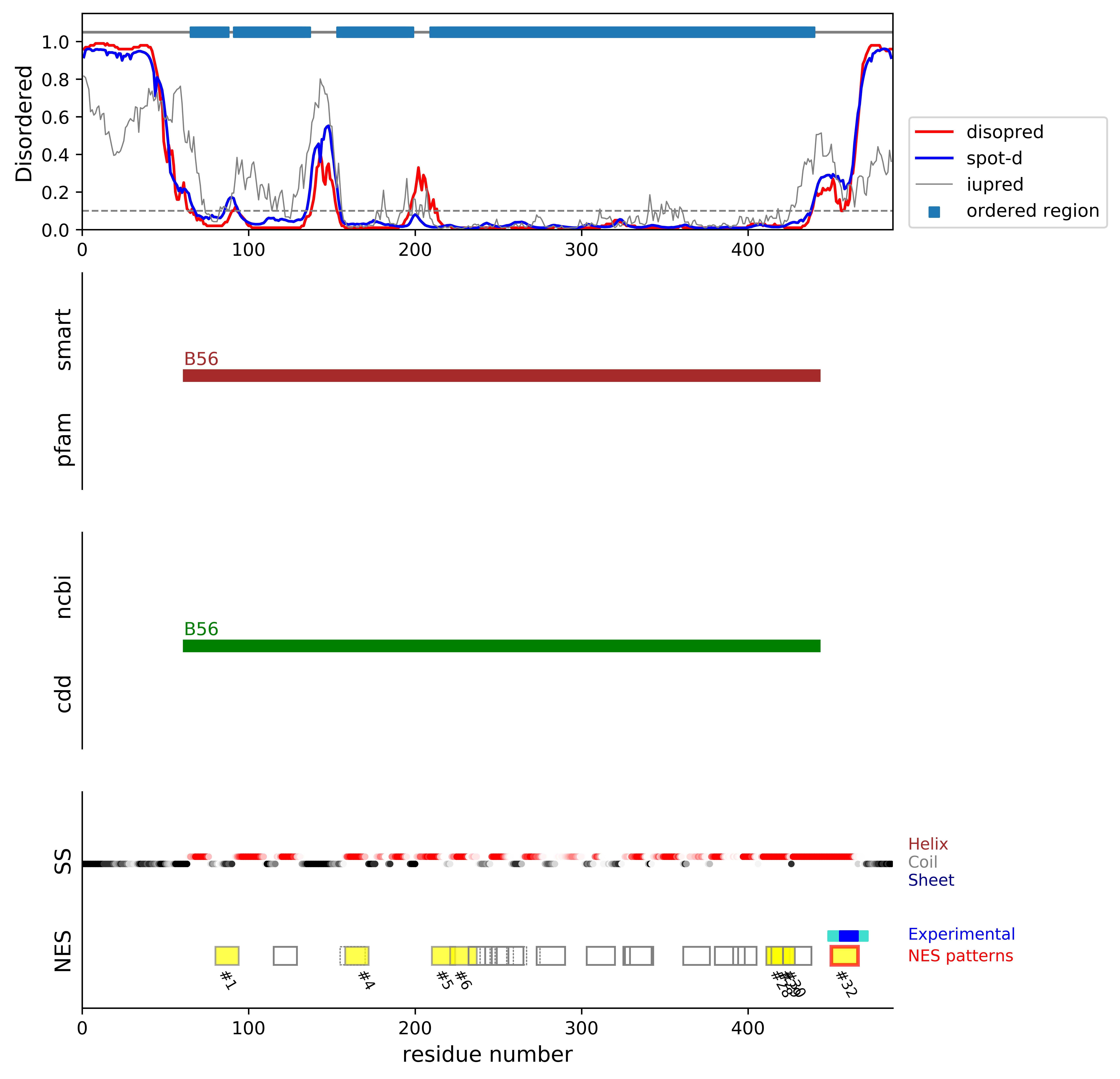

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q15172 | 80 | ILFDFMDSVSDLKS | CCCCCCCCCCCHHH | c3-4 | uniq | 0.055 | 0.114 | 0.145 | boundary | boundary|B56; | 0.0 |
| 2 | fp_D | Q15172 | 115 | IVESAYSDIVKMIS | CCHHHHHHHHHHHH | c3-AT | uniq | 0.01 | 0.05 | 0.134 | boundary | MID|B56; | 0.0 |
| 3 | fp_D | Q15172 | 155 | SWPHIQLVYEFFLRF | CCCHHHHHHHHHHHH | c1b-4 | multi | 0.013 | 0.036 | 0.039 | boundary | MID|B56; | 0.0 |
| 4 | fp_D | Q15172 | 158 | HIQLVYEFFLRFLE | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.01 | 0.029 | 0.018 | boundary | MID|B56; | 0.0 |
| 5 | fp_D | Q15172 | 210 | VLHRIYGKFLGLRA | HHHHHHHHCCCCHH | c3-4 | uniq | 0.052 | 0.017 | 0.017 | boundary | MID|B56; | 0.0 |
| 6 | fp_D | Q15172 | 221 | LRAFIRKQINNIFLRF | CHHHHHHHHHHHHHHH | c1a-4 | uniq | 0.011 | 0.011 | 0.013 | boundary | MID|B56; | 0.0 |
| 7 | fp_O | Q15172 | 232 | IFLRFIYETEHFNG | HHHHHHCCCCCCCC | c3-AT | uniq | 0.01 | 0.018 | 0.018 | ORD | MID|B56; | 0.0 |
| 8 | fp_O | Q15172 | 239 | ETEHFNGVAELLEILG | CCCCCCCHHHHHHHHH | c1a-AT-4 | multi | 0.01 | 0.022 | 0.018 | ORD | MID|B56; | 0.0 |
| 9 | fp_O | Q15172 | 239 | ETEHFNGVAELLEILG | CCCCCCCHHHHHHHHH | c1d-4 | multi | 0.01 | 0.022 | 0.018 | ORD | MID|B56; | 0.0 |
| 10 | fp_O | Q15172 | 242 | HFNGVAELLEILGS | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.011 | 0.022 | 0.018 | ORD | MID|B56; | 0.0 |
| 11 | fp_O | Q15172 | 245 | GVAELLEILGSIIN | CHHHHHHHHHHHHC | c3-4 | multi | 0.012 | 0.021 | 0.019 | ORD | MID|B56; | 0.0 |
| 12 | fp_O | Q15172 | 248 | ELLEILGSIINGFALPL | HHHHHHHHHHCCCCCCC | c1c-5 | multi | 0.012 | 0.027 | 0.021 | ORD | MID|B56; | 0.0 |
| 13 | fp_O | Q15172 | 249 | LLEILGSIINGFALPLKA | HHHHHHHHHCCCCCCCHH | c4-5 | multi | 0.012 | 0.028 | 0.02 | ORD | MID|B56; | 0.0 |
| 14 | fp_O | Q15172 | 249 | LLEILGSIINGFALPL | HHHHHHHHHCCCCCCC | c1a-5 | multi-selected | 0.012 | 0.027 | 0.02 | ORD | MID|B56; | 0.0 |
| 15 | fp_O | Q15172 | 249 | LLEILGSIINGFALPL | HHHHHHHHHCCCCCCC | c1d-5 | multi | 0.012 | 0.027 | 0.02 | ORD | MID|B56; | 0.0 |
| 16 | fp_O | Q15172 | 273 | MKVLIPMHTAKGLALFH | HHHHHCCCCCCCHHHHH | c1c-AT-4 | multi-selected | 0.011 | 0.016 | 0.028 | ORD | MID|B56; | 0.0 |
| 17 | fp_O | Q15172 | 275 | VLIPMHTAKGLALFH | HHHCCCCCCCHHHHH | c1b-AT-5 | multi | 0.011 | 0.017 | 0.022 | ORD | MID|B56; | 0.0 |
| 18 | fp_O | Q15172 | 303 | DTTLTEPVIRGLLKFWP | CCCCCHHHHHCCCCCCC | c1c-AT-4 | uniq | 0.018 | 0.019 | 0.047 | ORD | MID|B56; | 0.0 |
| 19 | fp_O | Q15172 | 325 | KEVMFLGEIEEILDVIE | HHHHHHHHHHHHHHHCC | c1c-4 | multi-selected | 0.016 | 0.019 | 0.054 | ORD | MID|B56; | 0.0 |
| 20 | fp_O | Q15172 | 325 | KEVMFLGEIEEILDVIE | HHHHHHHHHHHHHHHCC | c1cR-5 | multi | 0.016 | 0.019 | 0.054 | ORD | MID|B56; | 0.0 |
| 21 | fp_O | Q15172 | 326 | EVMFLGEIEEILDVIE | HHHHHHHHHHHHHHCC | c1aR-5 | multi-selected | 0.015 | 0.017 | 0.055 | ORD | MID|B56; | 0.0 |
| 22 | fp_O | Q15172 | 326 | EVMFLGEIEEILDVIE | HHHHHHHHHHHHHHCC | c1d-5 | multi | 0.015 | 0.017 | 0.055 | ORD | MID|B56; | 0.0 |
| 23 | fp_O | Q15172 | 329 | FLGEIEEILDVIEP | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.013 | 0.015 | 0.062 | ORD | MID|B56; | 0.0 |
| 24 | fp_O | Q15172 | 361 | SSSHFQVAERALYFWN | CCCHHHHHHHHHHHHC | c1d-AT-4 | uniq | 0.01 | 0.014 | 0.031 | ORD | MID|B56; | 0.0 |
| 25 | fp_O | Q15172 | 380 | ILSLIEENIDKILPIMFA | HHHHHHHHHHHHHHHHHH | c4-5 | multi-selected | 0.01 | 0.01 | 0.028 | ORD | MID|B56; | 0.0 |
| 26 | fp_O | Q15172 | 380 | ILSLIEENIDKILP | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.011 | 0.021 | ORD | MID|B56; | 0.0 |
| 27 | fp_O | Q15172 | 391 | ILPIMFASLYKISK | HHHHHHHHHHHHHH | c3-4 | uniq | 0.016 | 0.011 | 0.043 | ORD | MID|B56; | 0.0 |
| 28 | fp_D | Q15172 | 411 | IVALVYNVLKTLME | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.019 | 0.017 | 0.058 | boundary | boundary|B56; | 0.0 |
| 29 | fp_D | Q15172 | 411 | IVALVYNVLKTLMEMNG | HHHHHHHHHHHHHHHCH | c1c-5 | multi-selected | 0.017 | 0.021 | 0.068 | boundary | boundary|B56; | 0.0 |
| 30 | fp_D | Q15172 | 414 | LVYNVLKTLMEMNG | HHHHHHHHHHHHCH | c3-4 | multi-selected | 0.016 | 0.021 | 0.07 | boundary | boundary|B56; | 0.0 |
| 31 | fp_D | Q15172 | 421 | TLMEMNGKLFDDLTSSY | HHHHHCHHHHHHHHHHH | c1cR-4 | uniq | 0.018 | 0.044 | 0.19 | boundary | boundary|B56; | 0.0 |
| 32 | cand_D | Q15172 | 450 | EREELWKKLEELKLKK +++++++*++*+*++ | HHHHHHHHHHHHHHHH | c1a-4 | uniq | 0.212 | 0.297 | 0.242 | boundary | boundary|B56; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment