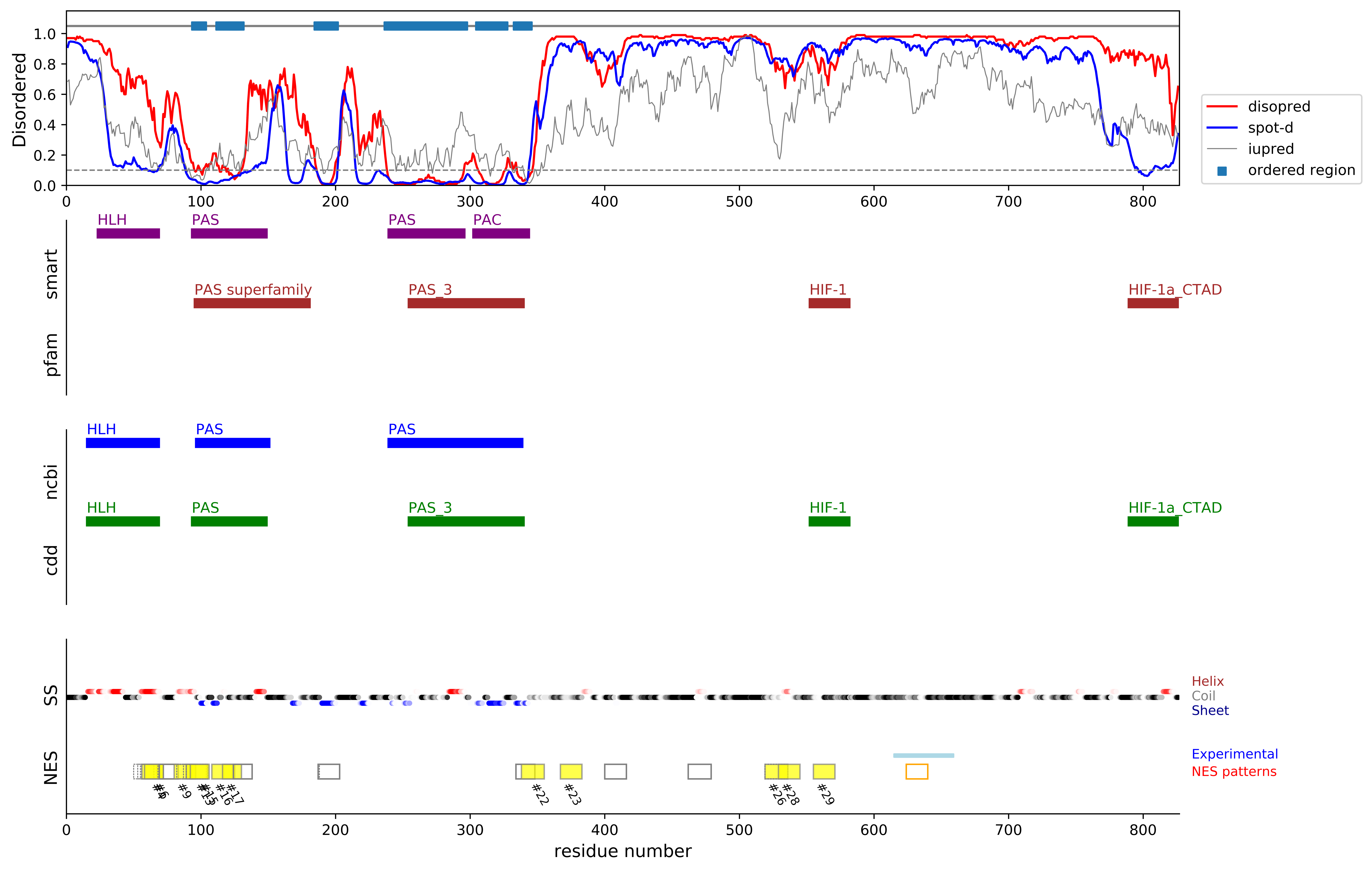

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q16665 | 50 | VSSHLDKASVMRLTI | CCCCCHHHHHHHHHH | c1b-AT-4 | multi | 0.633 | 0.118 | 0.28 | DISO | boundary|HLH; | 0.0 |
| 2 | fp_D | Q16665 | 53 | HLDKASVMRLTISY | CCHHHHHHHHHHHH | c2-AT-5 | multi | 0.597 | 0.104 | 0.233 | DISO | boundary|HLH; | 0.0 |
| 3 | fp_D | Q16665 | 55 | DKASVMRLTISYLRVRK | HHHHHHHHHHHHHHHCC | c1c-AT-4 | multi | 0.492 | 0.105 | 0.185 | DISO | boundary|HLH; | 0.0 |
| 4 | fp_D | Q16665 | 56 | KASVMRLTISYLRVRK | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.479 | 0.105 | 0.172 | DISO | boundary|HLH; | 0.0 |
| 5 | fp_D | Q16665 | 56 | KASVMRLTISYLRVRK | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.479 | 0.105 | 0.172 | DISO | boundary|HLH; | 0.0 |
| 6 | fp_D | Q16665 | 58 | SVMRLTISYLRVRK | HHHHHHHHHHHHCC | c2-5 | multi-selected | 0.448 | 0.105 | 0.157 | DISO | boundary|HLH; | 0.0 |
| 7 | fp_D | Q16665 | 68 | RVRKLLDAGDLDIED | HHCCCCCCCCCCCCC | c1b-AT-5 | multi | 0.467 | 0.264 | 0.203 | DISO | boundary|HLH; boundary|PAS; | 0.0 |
| 8 | fp_D | Q16665 | 69 | VRKLLDAGDLDIED | HCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.479 | 0.276 | 0.209 | DISO | boundary|HLH; boundary|PAS; | 0.0 |
| 9 | fp_D | Q16665 | 80 | IEDDMKAQMNCFYLKA | CCCHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.352 | 0.172 | 0.14 | DISO | boundary|HLH; boundary|PAS; | 0.0 |
| 10 | fp_D | Q16665 | 82 | DDMKAQMNCFYLKA | CHHHHHHHHHHHHH | c2-AT-4 | multi | 0.319 | 0.145 | 0.121 | DISO | boundary|HLH; boundary|PAS; | 0.0 |
| 11 | fp_D | Q16665 | 87 | QMNCFYLKALDGFVMVL | HHHHHHHHHCCCCEEEE | c1c-AT-5 | multi | 0.162 | 0.045 | 0.076 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 12 | fp_D | Q16665 | 89 | NCFYLKALDGFVMVL | HHHHHHHCCCCEEEE | c1b-4 | multi | 0.143 | 0.033 | 0.067 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 13 | fp_D | Q16665 | 89 | NCFYLKALDGFVMVLT | HHHHHHHCCCCEEEEE | c1aR-5 | multi-selected | 0.143 | 0.032 | 0.07 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 14 | fp_D | Q16665 | 89 | NCFYLKALDGFVMVLT | HHHHHHHCCCCEEEEE | c1d-4 | multi | 0.143 | 0.032 | 0.07 | boundary | boundary|HLH; boundary|PAS; | 0.0 |
| 15 | fp_D | Q16665 | 92 | YLKALDGFVMVLTD | HHHHCCCCEEEEEC | c3-4 | multi-selected | 0.119 | 0.023 | 0.065 | boundary | boundary|HLH; boundary|PAS; | 0.286 |
| 16 | fp_D | Q16665 | 108 | DMIYISDNVNKYMGLTQ | CEEEECCCHHHCCCCCC | c1c-5 | uniq | 0.122 | 0.031 | 0.172 | boundary | boundary|PAS; | 0.125 |
| 17 | fp_D | Q16665 | 116 | VNKYMGLTQFELTG | HHHCCCCCCCCCCC | c2-4 | uniq | 0.084 | 0.049 | 0.154 | boundary | boundary|PAS; | 0.0 |
| 18 | fp_D | Q16665 | 124 | QFELTGHSVFDFTH | CCCCCCCCCCCCCC | c3-AT | uniq | 0.229 | 0.077 | 0.193 | boundary | boundary|PAS; | 0.0 |
| 19 | fp_beta_D | Q16665 | 187 | ATWKVLHCTGHIHVYD | CCEEEEEEEEEEEECC | c1a-AT-4 | multi-selected | 0.054 | 0.031 | 0.175 | boundary | 1.0 | |
| 20 | fp_beta_D | Q16665 | 188 | TWKVLHCTGHIHVYD | CEEEEEEEEEEEECC | c1b-AT-4 | multi | 0.053 | 0.028 | 0.171 | boundary | 1.0 | |
| 21 | fp_beta_D | Q16665 | 334 | CIVCVNYVVSGIIQ | EEEEEEEEEECCCC | c3-4 | multi-selected | 0.067 | 0.092 | 0.073 | boundary | boundary|PAS_3; | 1.0 |
| 22 | fp_D | Q16665 | 338 | VNYVVSGIIQHDLIFSL | EEEEEECCCCCCCCCCC | c1c-4 | multi-selected | 0.261 | 0.261 | 0.072 | boundary | boundary|PAS_3; | 0.25 |
| 23 | fp_D | Q16665 | 367 | SDMKMTQLFTKVESED | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.969 | 0.925 | 0.396 | DISO | boundary|PAS_3; | 0.0 |
| 24 | fp_D | Q16665 | 400 | LAPAAGDTIISLDFGS | CCCCCCCCEECCCCCC | c1a-AT-4 | uniq | 0.812 | 0.79 | 0.429 | DISO | 0.286 | |
| 25 | fp_D | Q16665 | 462 | LRSSADPALNQEVALKL | CCCCCCCHHHHHHCCCC | c1c-AT-4 | uniq | 0.971 | 0.935 | 0.73 | DISO | 0.0 | |
| 26 | fp_D | Q16665 | 519 | YCFYVDSDMVNEFKLEL | CCCCCCCCCCCCCCHHH | c1c-4 | multi-selected | 0.818 | 0.836 | 0.386 | DISO | 0.0 | |
| 27 | fp_D | Q16665 | 519 | YCFYVDSDMVNEFKLEL | CCCCCCCCCCCCCCHHH | c1cR-4 | multi | 0.818 | 0.836 | 0.386 | DISO | 0.0 | |
| 28 | fp_D | Q16665 | 529 | NEFKLELVEKLFAEDT | CCCCHHHHHHHCCCCC | c1aR-4 | uniq | 0.751 | 0.8 | 0.418 | DISO | 0.0 | |
| 29 | fp_D | Q16665 | 555 | TDLDLEMLAPYIPMDD | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.789 | 0.889 | 0.564 | DISO | small|HIF-1; | 0.0 |
| 30 | fp_D | Q16665 | 624 | LKTVTKDRMEDIKILI ................ | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.983 | 0.903 | 0.578 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment