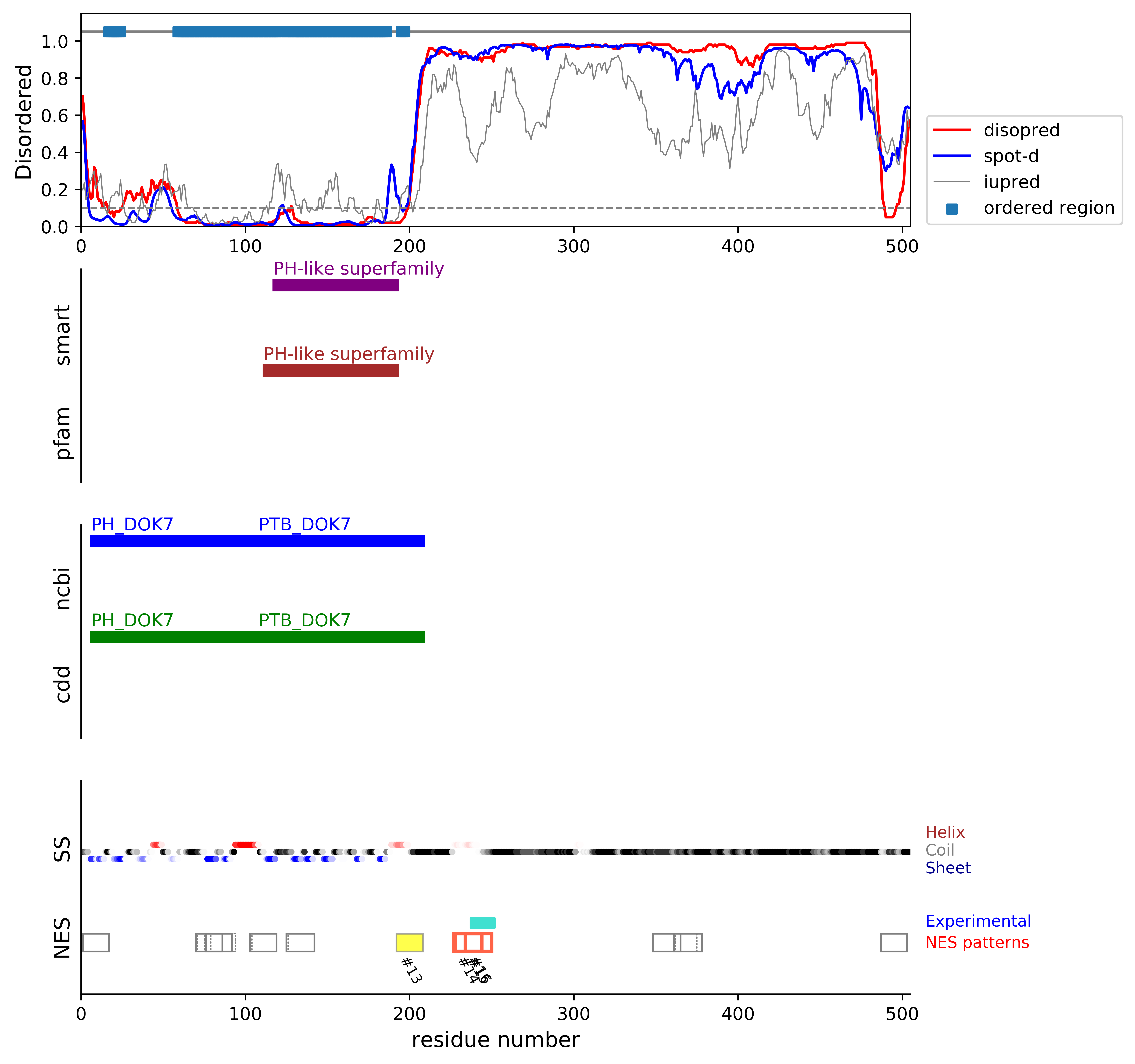

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q18PE1 | 1 | XMTEAALVEGQVKLRD | CCCCEEEEEEEEEECC | c1d-AT-5-Nt | uniq | 0.247 | 0.131 | 0.2 | boundary | boundary|PH_DOK7; | 1.0 |
| 2 | fp_beta_D | Q18PE1 | 70 | PYEGLVHTLAIVCLSQ | CCCCCCEEEEEEECCC | c1a-4 | multi-selected | 0.014 | 0.017 | 0.033 | boundary | MID|PH_DOK7; | 0.714 |
| 3 | fp_beta_D | Q18PE1 | 70 | PYEGLVHTLAIVCLSQ | CCCCCCEEEEEEECCC | c1d-AT-4 | multi | 0.014 | 0.017 | 0.033 | boundary | MID|PH_DOK7; | 0.714 |
| 4 | fp_beta_D | Q18PE1 | 71 | YEGLVHTLAIVCLSQ | CCCCCEEEEEEECCC | c1b-4 | multi | 0.013 | 0.015 | 0.03 | boundary | MID|PH_DOK7; | 0.857 |
| 5 | fp_beta_O | Q18PE1 | 75 | VHTLAIVCLSQAIMLGF | CEEEEEEECCCEEEEEE | c1c-AT-4 | multi | 0.012 | 0.009 | 0.026 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.625 |
| 6 | fp_beta_O | Q18PE1 | 76 | HTLAIVCLSQAIMLGF | EEEEEEECCCEEEEEE | c1aR-4 | multi-selected | 0.012 | 0.009 | 0.025 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.571 |
| 7 | fp_beta_O | Q18PE1 | 76 | HTLAIVCLSQAIMLGF | EEEEEEECCCEEEEEE | c1d-4 | multi | 0.012 | 0.009 | 0.025 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.571 |
| 8 | fp_beta_O | Q18PE1 | 79 | AIVCLSQAIMLGFDS | EEEECCCEEEEEECC | c1b-AT-5 | multi | 0.012 | 0.01 | 0.023 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.571 |
| 9 | fp_O | Q18PE1 | 103 | RIRYALGEVHRFHVTV | HHHHHHCCCEEEEEEE | c1a-AT-5 | multi-selected | 0.014 | 0.015 | 0.089 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.286 |
| 10 | fp_O | Q18PE1 | 104 | IRYALGEVHRFHVTV | HHHHHCCCEEEEEEE | c1b-4 | multi | 0.015 | 0.016 | 0.09 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.429 |
| 11 | fp_beta_O | Q18PE1 | 125 | ESGPATLHLCNDVLVLA | CCCCEEEEEECCEEEEE | c1c-AT-4 | multi-selected | 0.042 | 0.015 | 0.149 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.75 |
| 12 | fp_beta_O | Q18PE1 | 126 | SGPATLHLCNDVLVLA | CCCEEEEEECCEEEEE | c1d-AT-4 | multi | 0.039 | 0.012 | 0.143 | ORD | boundary|PH_DOK7; boundary|PTB_DOK7; | 0.714 |
| 13 | fp_D | Q18PE1 | 192 | ISFLFDCIVRGISPTK | HHHHHHHHCCCCCCCC | c1aR-4 | uniq | 0.253 | 0.322 | 0.154 | boundary | boundary|PTB_DOK7; | 0.0 |
| 14 | cand_D | Q18PE1 | 227 | VEERVAQEALETLQLEK ++++ | HHHHHHHHHHHHHHHCC | c1c-AT-4 | multi-selected | 0.924 | 0.918 | 0.553 | DISO | boundary|PTB_DOK7; | 0.0 |
| 15 | cand_D | Q18PE1 | 228 | EERVAQEALETLQLEK ++++ | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi-selected | 0.923 | 0.918 | 0.533 | DISO | boundary|PTB_DOK7; | 0.0 |
| 16 | cand_D | Q18PE1 | 234 | EALETLQLEKRLSLLS ++++++++++ | HHHHHHHHCCCCCCCC | c1d-AT-4 | uniq | 0.911 | 0.924 | 0.453 | DISO | boundary|PTB_DOK7; | 0.0 |
| 17 | fp_D | Q18PE1 | 348 | YSSSLSSYAGSSLDVWR | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.971 | 0.921 | 0.442 | DISO | 0.0 | |
| 18 | fp_D | Q18PE1 | 361 | DVWRATDELGSLLSLPA | CCCCCCCCCCCCCCCCC | c1c-AT-5 | multi-selected | 0.946 | 0.845 | 0.509 | DISO | 0.0 | |
| 19 | fp_D | Q18PE1 | 361 | DVWRATDELGSLLS | CCCCCCCCCCCCCC | c3-AT | multi | 0.946 | 0.864 | 0.488 | DISO | 0.0 | |
| 20 | fp_D | Q18PE1 | 362 | VWRATDELGSLLSLPA | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.946 | 0.842 | 0.512 | DISO | 0.0 | |
| 21 | fp_D | Q18PE1 | 487 | AFFSACPVCGGLKVNP | CCCCCCCCCCCCCCCC | c1d-AT-5 | uniq | 0.143 | 0.408 | 0.425 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment