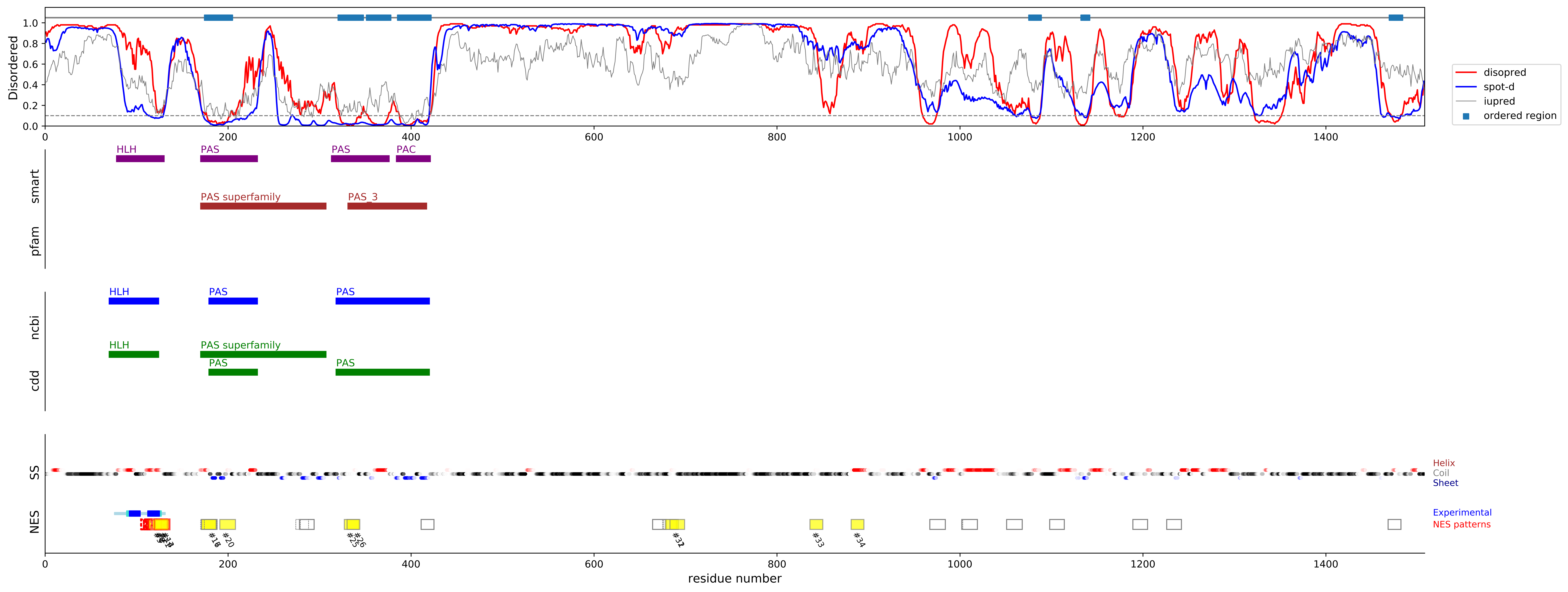

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | Q24167 | 105 | DVNQLDKASVMRITI ..........*+*++ | CCCCCHHHHHHHHHH | c1b-AT-5 | multi | 0.653 | 0.122 | 0.322 | DISO | boundary|HLH; | 0.0 |
| 2 | cand_D | Q24167 | 108 | QLDKASVMRITIAFLK .......*+*++++*+ | CCHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.529 | 0.097 | 0.257 | DISO | boundary|HLH; | 0.0 |
| 3 | cand_D | Q24167 | 108 | QLDKASVMRITIAF .......*+*++++ | CCHHHHHHHHHHHH | c2-AT-5 | multi | 0.584 | 0.1 | 0.271 | DISO | boundary|HLH; | 0.0 |
| 4 | cand_D | Q24167 | 110 | DKASVMRITIAFLKIRE .....*+*++++*++.. | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.435 | 0.089 | 0.218 | DISO | boundary|HLH; | 0.0 |
| 5 | cand_D | Q24167 | 111 | KASVMRITIAFLKIRE ....*+*++++*++.. | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.417 | 0.087 | 0.205 | DISO | boundary|HLH; | 0.0 |
| 6 | cand_D | Q24167 | 111 | KASVMRITIAFLKIRE ....*+*++++*++.. | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.417 | 0.087 | 0.205 | DISO | boundary|HLH; | 0.0 |
| 7 | cand_D | Q24167 | 113 | SVMRITIAFLKIRE ..*+*++++*++.. | HHHHHHHHHHHHHH | c2-5 | multi-selected | 0.375 | 0.084 | 0.19 | DISO | boundary|HLH; | 0.0 |
| 8 | cand_D | Q24167 | 113 | SVMRITIAFLKIRE ..*+*++++*++.. | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.375 | 0.084 | 0.19 | DISO | boundary|HLH; | 0.0 |
| 9 | cand_D | Q24167 | 116 | RITIAFLKIREMLQFVP +*++++*++...... | HHHHHHHHHHHHHCCCC | c1c-AT-5 | multi | 0.249 | 0.098 | 0.182 | DISO | boundary|HLH; | 0.0 |
| 10 | cand_D | Q24167 | 116 | RITIAFLKIREMLQ +*++++*++..... | HHHHHHHHHHHHHC | c3-AT | multi | 0.259 | 0.082 | 0.169 | DISO | boundary|HLH; | 0.0 |
| 11 | cand_D | Q24167 | 117 | ITIAFLKIREMLQFVP *++++*++...... | HHHHHHHHHHHHCCCC | c1aR-5 | multi-selected | 0.225 | 0.099 | 0.18 | DISO | boundary|HLH; | 0.0 |
| 12 | cand_D | Q24167 | 117 | ITIAFLKIREMLQFVP *++++*++...... | HHHHHHHHHHHHCCCC | c1d-4 | multi | 0.225 | 0.099 | 0.18 | DISO | boundary|HLH; | 0.0 |
| 13 | cand_D | Q24167 | 120 | AFLKIREMLQFVPSLR ++*++...... | HHHHHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.213 | 0.155 | 0.207 | DISO | boundary|HLH; | 0.0 |
| 14 | fp_D | Q24167 | 120 | AFLKIREMLQFVPS ++*++...... | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.186 | 0.117 | 0.185 | DISO | boundary|HLH; | 0.0 |
| 15 | fp_D | Q24167 | 170 | EARELLKQTMDGFLLVL | HHHHHHHHHCCCEEEEE | c1c-AT-4 | multi | 0.089 | 0.059 | 0.212 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.0 |
| 16 | fp_D | Q24167 | 171 | ARELLKQTMDGFLLVLS | HHHHHHHHCCCEEEEEC | c1c-4 | multi-selected | 0.079 | 0.047 | 0.2 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.125 |
| 17 | fp_D | Q24167 | 171 | ARELLKQTMDGFLLVL | HHHHHHHHCCCEEEEE | c1a-4 | multi-selected | 0.081 | 0.05 | 0.201 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.0 |
| 18 | fp_D | Q24167 | 171 | ARELLKQTMDGFLLVL | HHHHHHHHCCCEEEEE | c1d-AT-4 | multi | 0.081 | 0.05 | 0.201 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.0 |
| 19 | fp_D | Q24167 | 174 | LLKQTMDGFLLVLS | HHHHHCCCEEEEEC | c3-AT | multi | 0.066 | 0.031 | 0.168 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.286 |
| 20 | fp_D | Q24167 | 191 | DITYVSENVVEYLGITK | CEEEECCCHHHHCCCCC | c1c-5 | uniq | 0.07 | 0.033 | 0.13 | boundary | MID|PAS superfamily; boundary|PAS; | 0.125 |
| 21 | fp_beta_D | Q24167 | 274 | NIKSASYKVIHITG | CCCCCCEEEEEEEE | c3-AT | multi | 0.185 | 0.029 | 0.173 | DISO | boundary|PAS superfamily; | 0.571 |
| 22 | fp_beta_D | Q24167 | 278 | ASYKVIHITGHLVVNA | CCEEEEEEEEEEECCC | c1a-AT-4 | multi-selected | 0.19 | 0.018 | 0.144 | DISO | boundary|PAS superfamily; | 1.0 |
| 23 | fp_beta_D | Q24167 | 278 | ASYKVIHITGHLVVNA | CCEEEEEEEEEEECCC | c1d-4 | multi | 0.19 | 0.018 | 0.144 | DISO | boundary|PAS superfamily; | 1.0 |
| 24 | fp_beta_D | Q24167 | 279 | SYKVIHITGHLVVNA | CEEEEEEEEEEECCC | c1b-AT-4 | multi | 0.193 | 0.016 | 0.137 | DISO | boundary|PAS superfamily; | 1.0 |
| 25 | fp_D | Q24167 | 327 | LDMRFTYVDDKMHDLL | CCCCCCCCCCHHHHCC | c1aR-5 | multi-selected | 0.025 | 0.021 | 0.175 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.0 |
| 26 | fp_D | Q24167 | 330 | RFTYVDDKMHDLLG | CCCCCCCHHHHCCC | c3-4 | multi-selected | 0.031 | 0.02 | 0.175 | boundary | boundary|PAS superfamily; boundary|PAS; | 0.0 |
| 27 | fp_beta_D | Q24167 | 411 | SVVCVNYVISNLEN | EEEEEEEEECCCCC | c3-4 | uniq | 0.095 | 0.165 | 0.186 | boundary | boundary|PAS; | 0.857 |
| 28 | fp_D | Q24167 | 664 | LSHHLASTNCIQLDE | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.858 | 0.948 | 0.578 | DISO | 0.0 | |
| 29 | fp_D | Q24167 | 675 | QLDEMTPFSDMLVGL | CCCCCCCCCHHHHHC | c1b-5 | multi | 0.817 | 0.912 | 0.468 | DISO | 0.0 | |
| 30 | fp_D | Q24167 | 676 | LDEMTPFSDMLVGL | CCCCCCCCHHHHHC | c2-AT-4 | multi | 0.826 | 0.911 | 0.461 | DISO | 0.0 | |
| 31 | fp_D | Q24167 | 678 | EMTPFSDMLVGLMG | CCCCCCHHHHHCCC | c3-4 | multi-selected | 0.853 | 0.904 | 0.458 | DISO | 0.0 | |
| 32 | fp_D | Q24167 | 683 | SDMLVGLMGTCLLPED | CHHHHHCCCCCCCCCC | c1aR-4 | multi-selected | 0.911 | 0.889 | 0.449 | DISO | 0.0 | |
| 33 | fp_D | Q24167 | 836 | QEDDFSFEAFAMRA | CCCCCCHHHHCCCC | c2-4 | uniq | 0.731 | 0.774 | 0.593 | DISO | 0.0 | |
| 34 | fp_D | Q24167 | 881 | EIDAIQQQLQQLQQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.803 | 0.782 | 0.595 | DISO | 0.0 | |
| 35 | fp_beta_D | Q24167 | 967 | GNGTLSILAGSGVTVAE | CCCCEEEECCCCCCCHH | c1c-AT-4 | uniq | 0.237 | 0.222 | 0.372 | DISO | 0.5 | |
| 36 | fp_D | Q24167 | 1002 | NEFRTFQQLQQELQLQE | CCHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.669 | 0.271 | 0.49 | DISO | 0.0 | |
| 37 | fp_D | Q24167 | 1003 | EFRTFQQLQQELQLQE | CHHHHHHHHHHHHHHH | c1d-5 | multi | 0.666 | 0.269 | 0.485 | DISO | 0.0 | |
| 38 | fp_D | Q24167 | 1051 | YDVQMGGSLCHPMEDAF | CCCCCCCCCCCCCHHHH | c1cR-4 | uniq | 0.185 | 0.127 | 0.564 | boundary | 0.0 | |
| 39 | fp_D | Q24167 | 1098 | AASPTPCKVSAIQLLQ | CCCCCCCCCCHHHHHH | c1a-AT-4 | uniq | 0.773 | 0.536 | 0.492 | boundary | 0.0 | |
| 40 | fp_D | Q24167 | 1189 | PPTTTQSRMAKVNLVP | CCCCCCCCCCCCCCCH | c1a-AT-4 | uniq | 0.762 | 0.79 | 0.746 | DISO | 0.0 | |
| 41 | fp_D | Q24167 | 1226 | ESKRLKSGTLCLDVQS | CCCCCCCCCEEEECCC | c1a-AT-4 | uniq | 0.615 | 0.333 | 0.479 | DISO | 0.286 | |
| 42 | fp_D | Q24167 | 1468 | PLLHTPAMMDLVND | CCCCCHHHHHHCCC | c3-AT | uniq | 0.077 | 0.092 | 0.543 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment