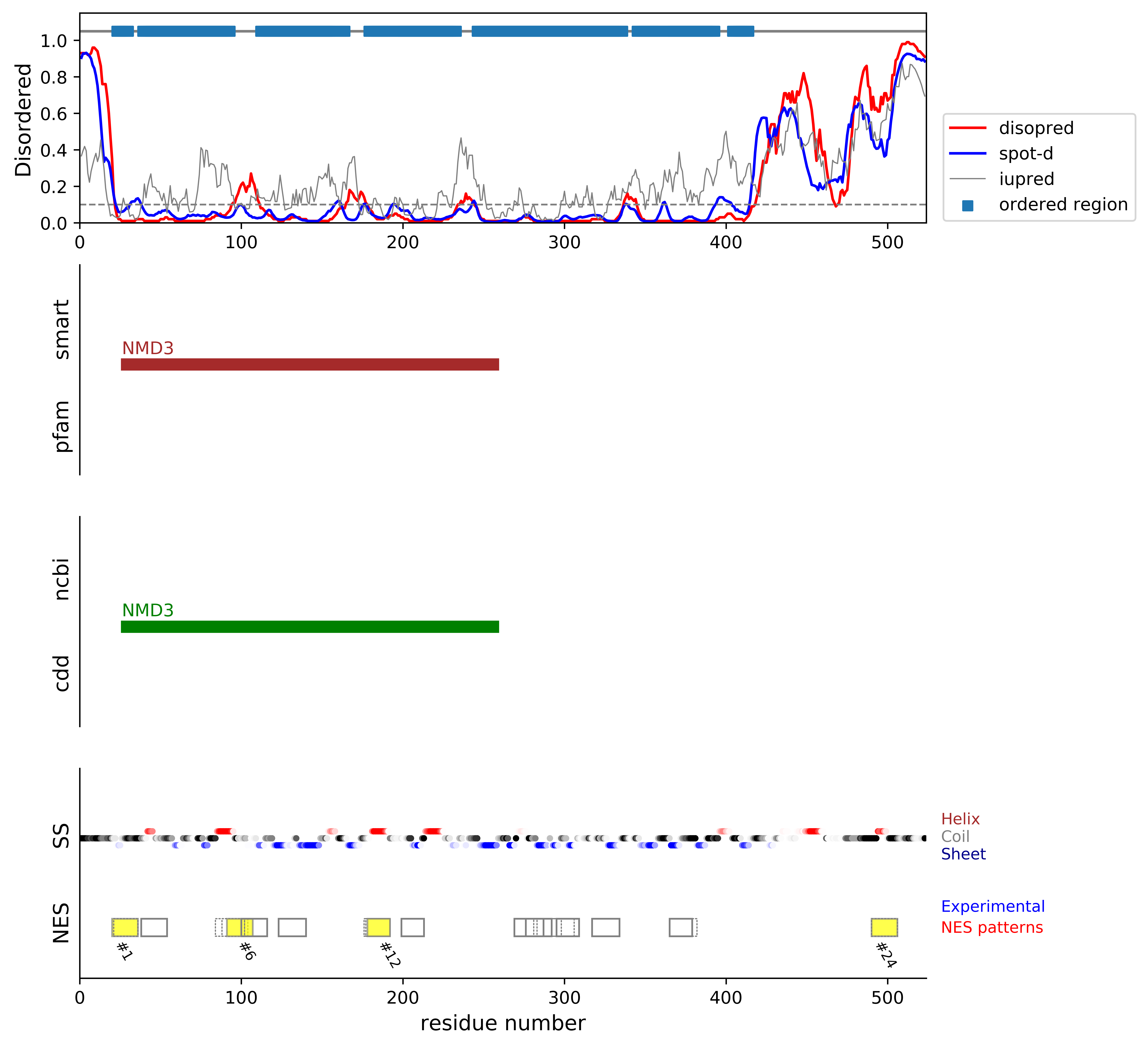

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q336P7 | 20 | TVGTVLCCICGVAMQP | CCCEEECCCCCCCCCC | c1a-5 | multi-selected | 0.049 | 0.101 | 0.054 | boundary | boundary|NMD3; | 0.286 |
| 2 | fp_D | Q336P7 | 21 | VGTVLCCICGVAMQP | CCEEECCCCCCCCCC | c1b-4 | multi | 0.029 | 0.093 | 0.054 | boundary | boundary|NMD3; | 0.143 |
| 3 | fp_D | Q336P7 | 38 | ANMCARCIRARVDITE | CCCCHHHHCCCCCCCC | c1d-AT-4 | uniq | 0.014 | 0.057 | 0.17 | boundary | boundary|NMD3; | 0.0 |
| 4 | fp_D | Q336P7 | 84 | SPELMQILLRRLNRPLAR | CHHHHHHHHHHHCCCCCC | c4-4 | multi | 0.095 | 0.054 | 0.193 | boundary | MID|NMD3; | 0.0 |
| 5 | fp_D | Q336P7 | 88 | MQILLRRLNRPLARLR | HHHHHHHHCCCCCCCC | c1aR-5 | multi | 0.119 | 0.057 | 0.172 | boundary | MID|NMD3; | 0.0 |
| 6 | fp_D | Q336P7 | 91 | LLRRLNRPLARLRVSL | HHHHHCCCCCCCCEEC | c1a-5 | multi-selected | 0.154 | 0.059 | 0.132 | boundary | MID|NMD3; | 0.0 |
| 7 | fp_D | Q336P7 | 100 | ARLRVSLSAAEFVFSE | CCCCEECCCCEEEEEC | c1a-AT-4 | multi-selected | 0.163 | 0.046 | 0.121 | boundary | MID|NMD3; | 0.429 |
| 8 | fp_D | Q336P7 | 102 | LRVSLSAAEFVFSE | CCEECCCCEEEEEC | c2-AT-4 | multi | 0.156 | 0.039 | 0.124 | boundary | MID|NMD3; | 0.429 |
| 9 | fp_beta_D | Q336P7 | 123 | LKLRLRREVFNAVVLEQ | EEEEEEEEEECCEEEEE | c1c-4 | uniq | 0.021 | 0.026 | 0.137 | __ | MID|NMD3; | 0.75 |
| 10 | fp_D | Q336P7 | 176 | PHRRTFLYLEQLLLKH | CCCCHHHHHHHHHHHH | c1a-AT-4 | multi | 0.051 | 0.051 | 0.074 | boundary | MID|NMD3; | 0.0 |
| 11 | fp_D | Q336P7 | 177 | HRRTFLYLEQLLLKH | CCCHHHHHHHHHHHH | c1b-4 | multi | 0.045 | 0.047 | 0.073 | boundary | MID|NMD3; | 0.0 |
| 12 | fp_D | Q336P7 | 178 | RRTFLYLEQLLLKH | CCHHHHHHHHHHHH | c2-4 | multi-selected | 0.041 | 0.043 | 0.072 | boundary | MID|NMD3; | 0.0 |
| 13 | fp_O | Q336P7 | 199 | RVAAAPGGLDFFFG | CCCCCCCEEEEEEC | c3-AT | uniq | 0.015 | 0.04 | 0.073 | ORD | MID|NMD3; | 0.429 |
| 14 | fp_O | Q336P7 | 269 | LSPQVSRDLGGLGPIVLC | ECHHHHHHCCCCCCEEEE | c4-4 | uniq | 0.018 | 0.023 | 0.079 | ORD | boundary|NMD3; | 0.0 |
| 15 | fp_beta_O | Q336P7 | 276 | DLGGLGPIVLCIKVTN | HCCCCCCEEEEEEECC | c1a-5 | multi-selected | 0.015 | 0.019 | 0.04 | ORD | boundary|NMD3; | 0.571 |
| 16 | fp_beta_O | Q336P7 | 276 | DLGGLGPIVLCIKVTN | HCCCCCCEEEEEEECC | c1d-5 | multi | 0.015 | 0.019 | 0.04 | ORD | boundary|NMD3; | 0.571 |
| 17 | fp_beta_O | Q336P7 | 281 | GPIVLCIKVTNAIALLD | CCEEEEEEECCEEEEEC | c1c-4 | multi | 0.01 | 0.01 | 0.032 | ORD | boundary|NMD3; | 0.75 |
| 18 | fp_beta_O | Q336P7 | 283 | IVLCIKVTNAIALLD | EEEEEEECCEEEEEC | c1b-AT-5 | multi | 0.01 | 0.009 | 0.026 | ORD | boundary|NMD3; | 0.714 |
| 19 | fp_O | Q336P7 | 292 | AIALLDPLTLRVHH | EEEEECCCCCEEEE | c3-AT | multi | 0.01 | 0.021 | 0.077 | ORD | boundary|NMD3; | 0.286 |
| 20 | fp_O | Q336P7 | 295 | LLDPLTLRVHHLEE | EECCCCCEEEEEEC | c3-4 | multi-selected | 0.01 | 0.024 | 0.1 | ORD | 0.429 | |
| 21 | fp_D | Q336P7 | 317 | KAALTSKQLVEYIVLDI | CCCCCCCCCCEEEEEEE | c1c-AT-4 | uniq | 0.016 | 0.024 | 0.092 | boundary | 0.25 | |
| 22 | fp_D | Q336P7 | 365 | TMFTVRTHLGHLLNPGD | CEEEEEECCCCCCCCCC | c1cR-4 | multi | 0.01 | 0.021 | 0.218 | boundary | 0.375 | |
| 23 | fp_beta_D | Q336P7 | 365 | TMFTVRTHLGHLLN | CEEEEEECCCCCCC | c3-4 | multi-selected | 0.01 | 0.019 | 0.235 | boundary | 0.571 | |
| 24 | fp_D | Q336P7 | 490 | PTVPIEELIEDLSLGD | CCCCHHHHHHCCCCCC | c1a-4 | multi-selected | 0.693 | 0.484 | 0.568 | DISO | 0.0 | |
| 25 | fp_D | Q336P7 | 490 | PTVPIEELIEDLSLGD | CCCCHHHHHHCCCCCC | c1d-4 | multi | 0.693 | 0.484 | 0.568 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment