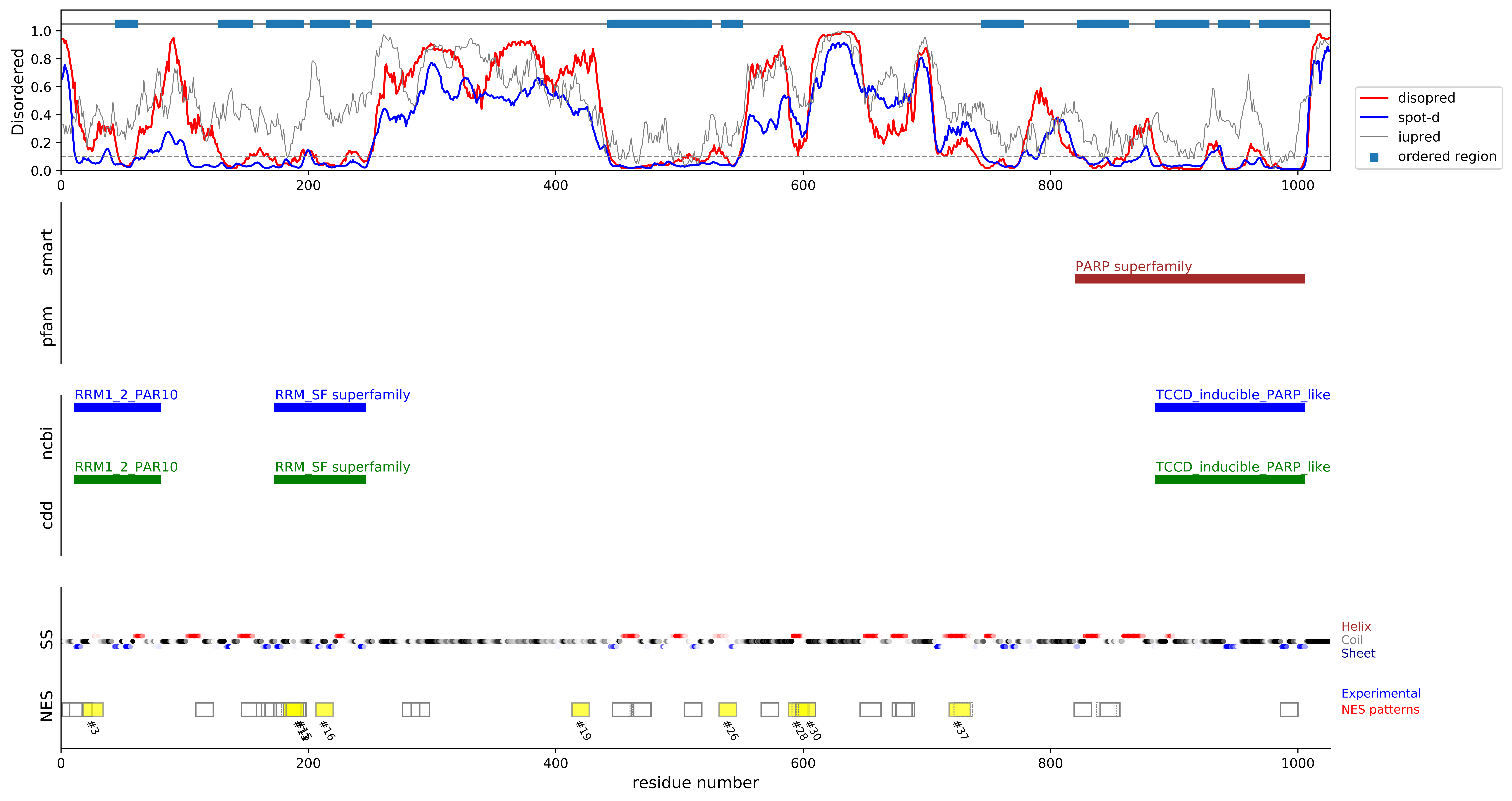

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q53GL7 | 1 | XMVAMAEAEAGVAVEV | CCCCCCCCCCCEEEEE | c1d-AT-5-Nt | uniq | 0.682 | 0.39 | 0.359 | DISO | boundary|RRM1_2_PAR10; | 0.0 |
| 2 | fp_beta_D | Q53GL7 | 7 | AEAGVAVEVRGLPPAVPD | CCCCCEEEEECCCCCCCC | c4-4 | uniq | 0.395 | 0.148 | 0.355 | DISO | boundary|RRM1_2_PAR10; | 0.625 |
| 3 | fp_D | Q53GL7 | 18 | LPPAVPDELLTLYFEN | CCCCCCCCHHHHHCCC | c1a-4 | uniq | 0.224 | 0.067 | 0.355 | boundary | boundary|RRM1_2_PAR10; | 0.0 |
| 4 | fp_D | Q53GL7 | 109 | VQALLRASGLPVQP | HHHHHHHCCCCCCC | c2-AT-4 | uniq | 0.291 | 0.031 | 0.333 | boundary | boundary|RRM1_2_PAR10; | 0.0 |
| 5 | fp_D | Q53GL7 | 146 | DVRVLEEQAQNLGLEG | HHHHHHHHHHCCCCCC | c1a-AT-5 | uniq | 0.099 | 0.04 | 0.402 | boundary | boundary|RRM_SF superfamily; | 0.0 |
| 6 | fp_beta_D | Q53GL7 | 158 | GLEGTLVSLARVPQ | CCCCCEEEEEECCC | c3-AT | uniq | 0.114 | 0.032 | 0.344 | boundary | boundary|RRM_SF superfamily; | 0.714 |
| 7 | fp_D | Q53GL7 | 165 | SLARVPQARAVRVVG | EEEECCCCCEEEEEC | c1b-AT-5 | uniq | 0.073 | 0.025 | 0.312 | __ | boundary|RRM_SF superfamily; | 0.286 |
| 8 | fp_D | Q53GL7 | 174 | AVRVVGDGASVDLLLLE | EEEEECCCCCCCCCCEE | c1c-AT-5 | multi-selected | 0.053 | 0.041 | 0.241 | boundary | boundary|RRM_SF superfamily; | 0.125 |
| 9 | fp_D | Q53GL7 | 178 | VGDGASVDLLLLEL | ECCCCCCCCCCEEE | c2-AT-4 | multi | 0.056 | 0.045 | 0.215 | boundary | boundary|RRM_SF superfamily; | 0.0 |
| 10 | fp_D | Q53GL7 | 178 | VGDGASVDLLLLELYL | ECCCCCCCCCCEEEEC | c1a-AT-4 | multi | 0.058 | 0.042 | 0.229 | boundary | boundary|RRM_SF superfamily; | 0.0 |
| 11 | fp_D | Q53GL7 | 180 | DGASVDLLLLELYLEN | CCCCCCCCCEEEECCC | c1a-4 | multi-selected | 0.07 | 0.042 | 0.238 | boundary | boundary|RRM_SF superfamily; | 0.286 |
| 12 | fp_D | Q53GL7 | 180 | DGASVDLLLLELYLEN | CCCCCCCCCEEEECCC | c1d-4 | multi | 0.07 | 0.042 | 0.238 | boundary | boundary|RRM_SF superfamily; | 0.286 |
| 13 | fp_D | Q53GL7 | 180 | DGASVDLLLLELYL | CCCCCCCCCEEEEC | c2-4 | multi-selected | 0.061 | 0.043 | 0.226 | boundary | boundary|RRM_SF superfamily; | 0.143 |
| 14 | fp_beta_D | Q53GL7 | 182 | ASVDLLLLELYLENER | CCCCCCCEEEECCCCC | c1aR-4 | multi-selected | 0.08 | 0.045 | 0.253 | boundary | boundary|RRM_SF superfamily; | 0.571 |
| 15 | fp_D | Q53GL7 | 182 | ASVDLLLLELYLEN | CCCCCCCEEEECCC | c2-4 | multi-selected | 0.073 | 0.039 | 0.237 | boundary | boundary|RRM_SF superfamily; | 0.429 |
| 16 | fp_D | Q53GL7 | 206 | DLQRLPGPLGTVAS | CEEEECCCCEEEEE | c3-4 | uniq | 0.051 | 0.039 | 0.507 | boundary | MID|RRM_SF superfamily; | 0.429 |
| 17 | fp_D | Q53GL7 | 276 | ALLRTGGLVTALQG | CCCCCCCCCCCCCC | c3-AT | uniq | 0.709 | 0.412 | 0.537 | DISO | boundary|RRM_SF superfamily; | 0.0 |
| 18 | fp_D | Q53GL7 | 283 | LVTALQGAGTVTMGS | CCCCCCCCCCCCCCC | c1b-AT-5 | uniq | 0.806 | 0.535 | 0.661 | DISO | 0.0 | |
| 19 | fp_D | Q53GL7 | 413 | LVGPMEITMGSLEK | CCCCCCCCCCCCCC | c3-4 | uniq | 0.774 | 0.428 | 0.56 | DISO | 0.0 | |
| 20 | fp_D | Q53GL7 | 446 | MVLLMEPGAMRFLQ | EEEECCCHHHHHHH | c3-AT | multi | 0.034 | 0.029 | 0.14 | boundary | 0.143 | |
| 21 | fp_D | Q53GL7 | 446 | MVLLMEPGAMRFLQLYH | EEEECCCHHHHHHHHHH | c1c-AT-5 | multi-selected | 0.031 | 0.028 | 0.149 | boundary | 0.0 | |
| 22 | fp_O | Q53GL7 | 461 | YHEDLLAGLGDVALLP | HHHHHHCCCCCCEEEC | c1a-4 | multi-selected | 0.025 | 0.021 | 0.184 | ORD | 0.0 | |
| 23 | fp_O | Q53GL7 | 462 | HEDLLAGLGDVALLP | HHHHHCCCCCCEEEC | c1b-4 | multi | 0.025 | 0.021 | 0.181 | ORD | 0.0 | |
| 24 | fp_O | Q53GL7 | 463 | EDLLAGLGDVALLP | HHHHCCCCCCEEEC | c2-AT-4 | multi | 0.026 | 0.022 | 0.183 | ORD | 0.0 | |
| 25 | fp_beta_D | Q53GL7 | 504 | LLGSISCHVLCLEH | HHCCCCEEEEEECC | c3-4 | uniq | 0.089 | 0.037 | 0.068 | boundary | 0.571 | |
| 26 | fp_D | Q53GL7 | 532 | LLQGLEAQFQCVFG | HHHHHHHCEEEEEE | c3-4 | uniq | 0.084 | 0.038 | 0.23 | boundary | 0.286 | |
| 27 | fp_D | Q53GL7 | 566 | ALPVLPGNAHTLWT | CCCCCCCCCCCCCC | c3-AT | uniq | 0.754 | 0.315 | 0.683 | DISO | 0.0 | |
| 28 | fp_D | Q53GL7 | 588 | EDVSLEEVRELLATLE | CCCCHHHHHHHHHHCC | c1aR-5 | multi-selected | 0.237 | 0.353 | 0.564 | DISO | 0.0 | |
| 29 | fp_D | Q53GL7 | 591 | SLEEVRELLATLEG | CHHHHHHHHHHCCC | c3-4 | multi | 0.207 | 0.325 | 0.523 | DISO | 0.0 | |
| 30 | fp_D | Q53GL7 | 594 | EVRELLATLEGLDLDG | HHHHHHHHCCCCCCCC | c1a-5 | multi-selected | 0.357 | 0.373 | 0.5 | DISO | 0.0 | |
| 31 | fp_D | Q53GL7 | 594 | EVRELLATLEGLDLDG | HHHHHHHHCCCCCCCC | c1d-AT-5 | multi | 0.357 | 0.373 | 0.5 | DISO | 0.0 | |
| 32 | fp_D | Q53GL7 | 595 | VRELLATLEGLDLDG | HHHHHHHCCCCCCCC | c1b-4 | multi | 0.368 | 0.373 | 0.493 | DISO | 0.0 | |
| 33 | fp_D | Q53GL7 | 596 | RELLATLEGLDLDG | HHHHHHCCCCCCCC | c2-AT-4 | multi | 0.383 | 0.376 | 0.491 | DISO | 0.0 | |
| 34 | fp_D | Q53GL7 | 646 | APRWLEEEAALQLALHR | CCCCHHHHHHHHHHHHH | c1c-AT-4 | uniq | 0.452 | 0.566 | 0.564 | DISO | 0.0 | |
| 35 | fp_D | Q53GL7 | 672 | EQEEAAALRQALTLSL | HHHHHHHHHHHHCCCC | c1d-AT-4 | multi-selected | 0.334 | 0.502 | 0.565 | DISO | 0.0 | |
| 36 | fp_D | Q53GL7 | 675 | EAAALRQALTLSLLE | HHHHHHHHHCCCCCC | c1b-AT-4 | multi-selected | 0.342 | 0.521 | 0.545 | DISO | 0.0 | |
| 37 | fp_D | Q53GL7 | 718 | DVEELDRALRAALEVHV | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.186 | 0.303 | 0.411 | boundary | 0.0 | |
| 38 | fp_D | Q53GL7 | 722 | LDRALRAALEVHVQE | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.193 | 0.302 | 0.418 | boundary | 0.0 | |
| 39 | fp_D | Q53GL7 | 819 | NLERLAENTGEFQE | CEEECCCCCHHHHH | c3-AT | uniq | 0.087 | 0.088 | 0.314 | boundary | 0.143 | |
| 40 | fp_D | Q53GL7 | 837 | FYDTLDAARSSIRVVR | HHHHHHHCCCCCCCCC | c1d-AT-4 | multi | 0.041 | 0.062 | 0.173 | boundary | 0.0 | |
| 41 | fp_D | Q53GL7 | 840 | TLDAARSSIRVVRVER | HHHHCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.052 | 0.059 | 0.19 | boundary | 0.0 | |
| 42 | fp_D | Q53GL7 | 986 | SIFVIFHDTQALPT | CEEEEECCCCCCCC | c3-AT | uniq | 0.011 | 0.009 | 0.113 | boundary | boundary|TCCD_inducible_PARP_like; | 0.429 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment