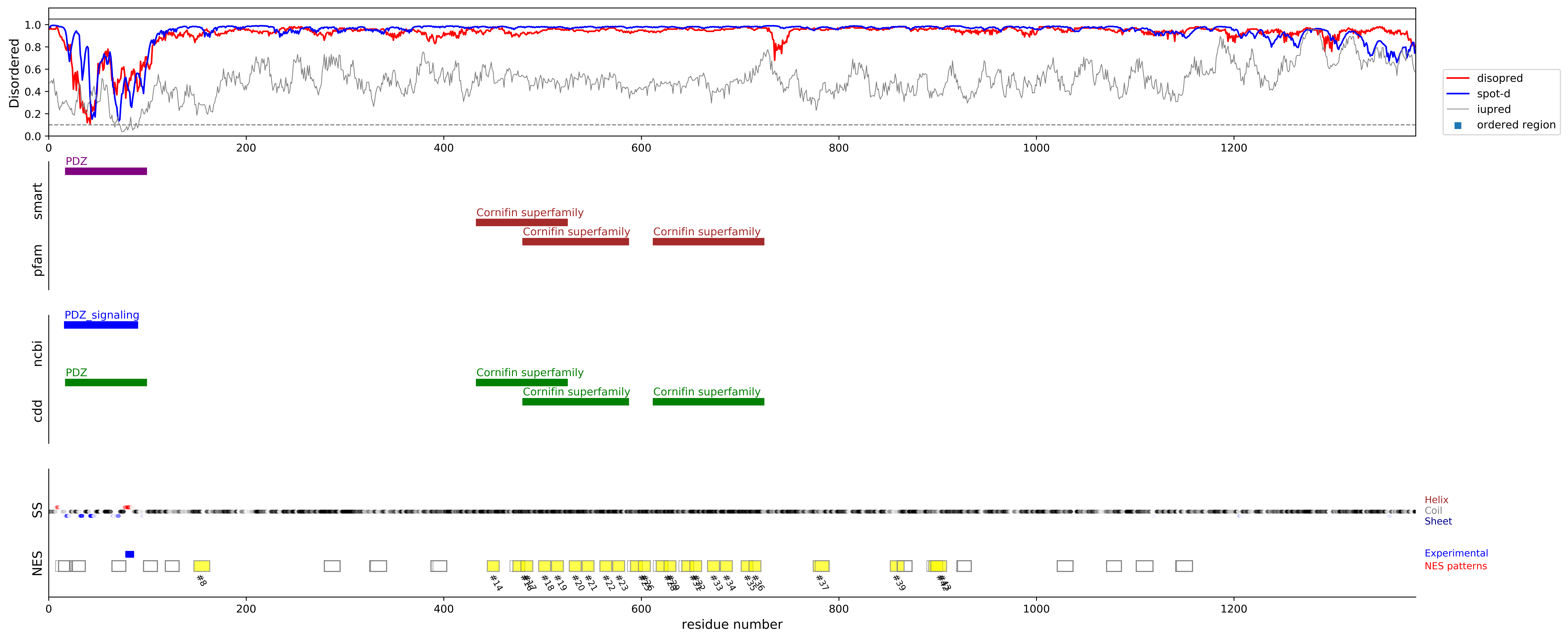

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q63425 | 7 | SAEELRRAELVEIIV | CHHHHHCCCCEEEEE | c1b-AT-4 | multi | 0.87 | 0.925 | 0.299 | DISO | boundary|PDZ; | 0.143 |
| 2 | fp_D | Q63425 | 10 | ELRRAELVEIIVET | HHHCCCCEEEEECC | c2-AT-5 | multi-selected | 0.831 | 0.889 | 0.276 | DISO | boundary|PDZ; | 0.429 |
| 3 | fp_D | Q63425 | 21 | VETEAQTGVSGFNVAG | ECCCCCCCCCCEEECC | c1a-AT-4 | multi-selected | 0.477 | 0.788 | 0.294 | DISO | boundary|PDZ; | 0.0 |
| 4 | fp_D | Q63425 | 23 | TEAQTGVSGFNVAG | CCCCCCCCCEEECC | c2-AT-4 | multi | 0.432 | 0.8 | 0.301 | DISO | boundary|PDZ; | 0.143 |
| 5 | fp_beta_D | Q63425 | 64 | QLLSARVFFENFKY | EEEEEEEECCCCCH | c3-AT | uniq | 0.504 | 0.344 | 0.101 | DISO | boundary|PDZ; | 0.714 |
| 6 | fp_D | Q63425 | 96 | LKRTVPTGDLALRP | EECCCCCCCCCCCC | c2-AT-4 | uniq | 0.716 | 0.734 | 0.31 | DISO | boundary|PDZ; | 0.0 |
| 7 | fp_D | Q63425 | 118 | KGPRAKVAKLNIQS | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.908 | 0.951 | 0.394 | DISO | boundary|PDZ; | 0.0 |
| 8 | fp_D | Q63425 | 147 | TPADLAPVDVEFSFPK | CCCCCCCCCCCCCCCC | c1d-4 | uniq | 0.911 | 0.954 | 0.264 | DISO | 0.0 | |
| 9 | fp_D | Q63425 | 279 | AAPAVEPPTTGIQVPQ | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.927 | 0.967 | 0.668 | DISO | 0.0 | |
| 10 | fp_D | Q63425 | 325 | TLDVAAPSVEVDLALPG | CCCCCCCCCCCCCCCCC | c1c-AT-5 | multi-selected | 0.955 | 0.942 | 0.419 | DISO | 0.0 | |
| 11 | fp_D | Q63425 | 326 | LDVAAPSVEVDLALPG | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.955 | 0.942 | 0.419 | DISO | 0.0 | |
| 12 | fp_D | Q63425 | 387 | KGPRLRMPTFGLSLLE | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.888 | 0.98 | 0.542 | DISO | 0.0 | |
| 13 | fp_D | Q63425 | 389 | PRLRMPTFGLSLLE | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.886 | 0.98 | 0.538 | DISO | 0.0 | |
| 14 | fp_D | Q63425 | 444 | KLPKVPEIKLPK | CCCCCCCCCCCC | c2-rev | uniq | 0.963 | 0.976 | 0.542 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 15 | fp_D | Q63425 | 467 | PEVQLPKMSDMKLPK | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.912 | 0.973 | 0.547 | DISO | MID|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 16 | fp_D | Q63425 | 470 | QLPKMSDMKLPK | CCCCCCCCCCCC | c2-rev | multi-selected | 0.912 | 0.974 | 0.546 | DISO | MID|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 17 | fp_D | Q63425 | 478 | KLPKIPEMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.922 | 0.979 | 0.504 | DISO | MID|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 18 | fp_D | Q63425 | 496 | KLPKVPEMKVPE | CCCCCCCCCCCC | c2-rev | uniq | 0.948 | 0.979 | 0.495 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 19 | fp_D | Q63425 | 509 | KLPKIPEMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.961 | 0.977 | 0.469 | DISO | boundary|Cornifin superfamily; MID|Cornifin superfamily; | 0.0 |
| 20 | fp_D | Q63425 | 527 | QLPKVPEMKLPD | CCCCCCCCCCCC | c2-rev | uniq | 0.951 | 0.97 | 0.516 | DISO | boundary|Cornifin superfamily; MID|Cornifin superfamily; | 0.0 |
| 21 | fp_D | Q63425 | 540 | KLPKVPEMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.941 | 0.98 | 0.505 | DISO | boundary|Cornifin superfamily; MID|Cornifin superfamily; | 0.0 |
| 22 | fp_D | Q63425 | 558 | QLPKVPEMKLPD | CCCCCCCCCCCC | c2-rev | uniq | 0.962 | 0.979 | 0.514 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 23 | fp_D | Q63425 | 571 | KLPKVPEMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.952 | 0.982 | 0.481 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 24 | fp_D | Q63425 | 586 | PEVQLPKVSEVKLPK | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.939 | 0.977 | 0.404 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 25 | fp_D | Q63425 | 589 | QLPKVSEVKLPK | CCCCCCCCCCCC | c2-rev | multi-selected | 0.94 | 0.978 | 0.406 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 26 | fp_D | Q63425 | 597 | KLPKIPDMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.945 | 0.98 | 0.428 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 27 | fp_D | Q63425 | 612 | PELQLPKMSEVKLPK | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.942 | 0.979 | 0.468 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 28 | fp_D | Q63425 | 615 | QLPKMSEVKLPK | CCCCCCCCCCCC | c2-rev | multi-selected | 0.944 | 0.98 | 0.467 | DISO | boundary|Cornifin superfamily; boundary|Cornifin superfamily; | 0.0 |
| 29 | fp_D | Q63425 | 623 | KLPKIPDMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.958 | 0.979 | 0.465 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 30 | fp_D | Q63425 | 638 | PEVQLPKVSELKLPK | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.965 | 0.975 | 0.474 | DISO | MID|Cornifin superfamily; | 0.0 |
| 31 | fp_D | Q63425 | 641 | QLPKVSELKLPK | CCCCCCCCCCCC | c2-rev | multi-selected | 0.964 | 0.976 | 0.48 | DISO | MID|Cornifin superfamily; | 0.0 |
| 32 | fp_D | Q63425 | 649 | KLPKVPEMTMPD | CCCCCCCCCCCC | c2-rev | uniq | 0.957 | 0.981 | 0.515 | DISO | MID|Cornifin superfamily; | 0.0 |
| 33 | fp_D | Q63425 | 667 | QLPKVPDIKLPE | CCCCCCCCCCCC | c2-rev | uniq | 0.944 | 0.979 | 0.404 | DISO | MID|Cornifin superfamily; | 0.0 |
| 34 | fp_D | Q63425 | 680 | KLPKVPEMAVPD | CCCCCCCCCCCC | c2-rev | uniq | 0.958 | 0.981 | 0.432 | DISO | MID|Cornifin superfamily; | 0.0 |
| 35 | fp_D | Q63425 | 701 | KVPQVPDVHLPK | CCCCCCCCCCCC | c2-rev | uniq | 0.975 | 0.982 | 0.506 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 36 | fp_D | Q63425 | 709 | HLPKVPEMKLPK | CCCCCCCCCCCC | c2-rev | uniq | 0.963 | 0.982 | 0.569 | DISO | boundary|Cornifin superfamily; | 0.0 |
| 37 | fp_D | Q63425 | 774 | LQPEVGTEVARVGVPS | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.957 | 0.97 | 0.374 | DISO | 0.0 | |
| 38 | fp_D | Q63425 | 776 | PEVGTEVARVGVPS | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.956 | 0.973 | 0.38 | DISO | 0.0 | |
| 39 | fp_D | Q63425 | 852 | KKEQLEMVEMKVKP | CCCCCCCCCCCCCC | c2-4 | uniq | 0.978 | 0.98 | 0.464 | DISO | 0.0 | |
| 40 | fp_D | Q63425 | 859 | VEMKVKPTSKFSLPK | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.973 | 0.976 | 0.411 | DISO | 0.0 | |
| 41 | fp_D | Q63425 | 889 | PGRATKLKVSKFAISL | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi | 0.965 | 0.979 | 0.394 | DISO | 0.0 | |
| 42 | fp_D | Q63425 | 891 | RATKLKVSKFAISL | CCCCCCCCCCCCCC | c2-4 | multi-selected | 0.966 | 0.978 | 0.382 | DISO | 0.0 | |
| 43 | fp_D | Q63425 | 893 | TKLKVSKFAISLPRAR | CCCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.966 | 0.976 | 0.394 | DISO | 0.0 | |
| 44 | fp_D | Q63425 | 919 | GEAGLLPALDLSIPQ | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.939 | 0.979 | 0.383 | DISO | 0.0 | |
| 45 | fp_D | Q63425 | 920 | EAGLLPALDLSIPQ | CCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.939 | 0.978 | 0.376 | DISO | 0.0 | |
| 46 | fp_D | Q63425 | 1021 | PGEKLEAIAGQLKIPE | CCCCCCCCCCCCCCCE | c1a-AT-4 | multi-selected | 0.948 | 0.978 | 0.451 | DISO | 0.0 | |
| 47 | fp_D | Q63425 | 1021 | PGEKLEAIAGQLKIPE | CCCCCCCCCCCCCCCE | c1d-4 | multi | 0.948 | 0.978 | 0.451 | DISO | 0.0 | |
| 48 | fp_D | Q63425 | 1071 | GQEGAQRVSSLGISL | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.947 | 0.98 | 0.448 | DISO | 0.0 | |
| 49 | fp_D | Q63425 | 1101 | AAPSAEGTVGSRIQVPQ | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.936 | 0.958 | 0.566 | DISO | 0.0 | |
| 50 | fp_D | Q63425 | 1141 | MPTVTVPQLELDVGLGH | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.942 | 0.911 | 0.45 | DISO | 0.0 | |
| 51 | fp_D | Q63425 | 1142 | PTVTVPQLELDVGLGH | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.943 | 0.909 | 0.459 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment