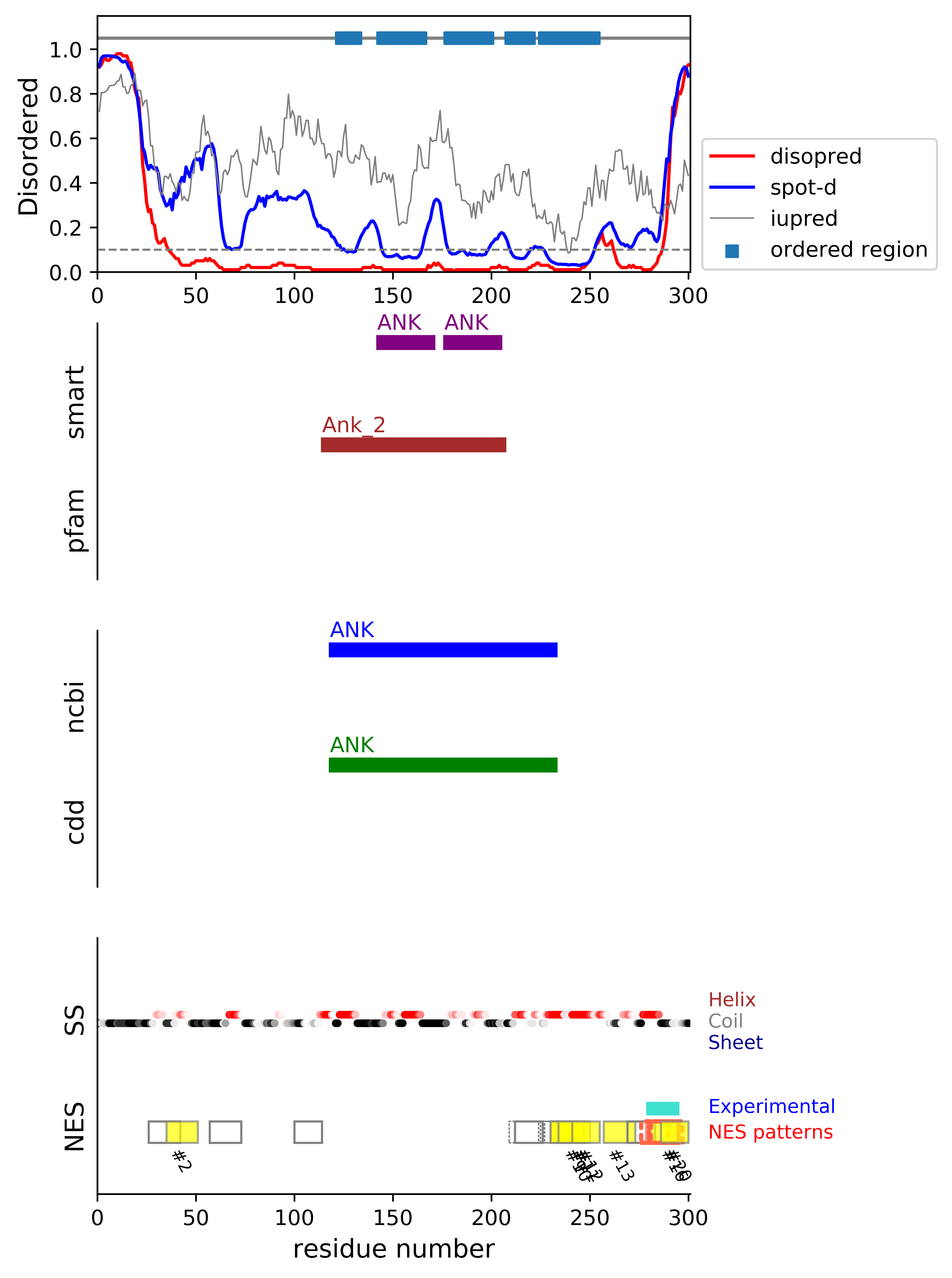

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q6NXT1 | 26 | APEPLTDAEGLFSFAD | CCCHHHHHCCCCCCHH | c1d-AT-4 | uniq | 0.141 | 0.378 | 0.448 | DISO | 0.0 | |
| 2 | fp_D | Q6NXT1 | 35 | GLFSFADFGSALGGGG | CCCCCHHHHHHHCCCC | c1aR-4 | uniq | 0.056 | 0.393 | 0.395 | DISO | 0.0 | |
| 3 | fp_D | Q6NXT1 | 57 | ASGGAQSPLRYLHVLW | CCCCCCCCCHHHHHHH | c1a-AT-4 | uniq | 0.023 | 0.258 | 0.479 | DISO | 0.0 | |
| 4 | fp_D | Q6NXT1 | 100 | RLGPTGKEVHALKR | CCCCCCCCCCCCCH | c3-AT | uniq | 0.017 | 0.304 | 0.641 | boundary | boundary|ANK; | 0.0 |
| 5 | fp_D | Q6NXT1 | 209 | GRTPLHLAKSKLNILQ | CCCHHHHHHHHCCHHH | c1d-AT-4 | multi | 0.017 | 0.084 | 0.375 | boundary | boundary|ANK; | 0.0 |
| 6 | fp_D | Q6NXT1 | 212 | PLHLAKSKLNILQE | HHHHHHHHCCHHHH | c3-AT | multi-selected | 0.02 | 0.085 | 0.351 | boundary | boundary|ANK; | 0.0 |
| 7 | fp_D | Q6NXT1 | 224 | QEGHAQCLEAVRLEV | HHCCHHHHHHHHHHH | c1b-AT-4 | multi | 0.02 | 0.06 | 0.21 | boundary | boundary|ANK; | 0.0 |
| 8 | fp_D | Q6NXT1 | 227 | HAQCLEAVRLEVKQ | CHHHHHHHHHHHHH | c2-AT-4 | multi | 0.016 | 0.046 | 0.185 | boundary | boundary|ANK; | 0.0 |
| 9 | fp_D | Q6NXT1 | 230 | CLEAVRLEVKQIIH | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.011 | 0.036 | 0.166 | boundary | boundary|ANK; | 0.0 |
| 10 | fp_D | Q6NXT1 | 230 | CLEAVRLEVKQIIHMLR | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.012 | 0.035 | 0.173 | boundary | boundary|ANK; | 0.0 |
| 11 | fp_D | Q6NXT1 | 234 | VRLEVKQIIHMLREYL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.014 | 0.034 | 0.197 | boundary | boundary|ANK; | 0.0 |
| 12 | fp_D | Q6NXT1 | 241 | IIHMLREYLERLGQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.044 | 0.056 | 0.286 | boundary | boundary|ANK; | 0.0 |
| 13 | fp_D | Q6NXT1 | 257 | QRERLDDLCTRLQMTS | HHHCCCCCCCHHHHHH | c1d-4 | uniq | 0.066 | 0.159 | 0.46 | boundary | boundary|ANK; | 0.0 |
| 14 | fp_D | Q6NXT1 | 269 | QMTSTKEQVDEVTD + | HHHHHCCCHHHHHH | c3-AT | uniq | 0.018 | 0.158 | 0.387 | DISO | 0.0 | |
| 15 | cand_D | Q6NXT1 | 276 | QVDEVTDLLASFTS ++++++++ | CHHHHHHHHHCCCC | c3-4 | multi | 0.058 | 0.227 | 0.321 | DISO | 0.0 | |
| 16 | cand_D | Q6NXT1 | 279 | EVTDLLASFTSLSLQM +++++++++++ | HHHHHHHCCCCCCCCC | c1a-5 | multi-selected | 0.247 | 0.372 | 0.302 | DISO | 0.0 | |
| 17 | cand_D | Q6NXT1 | 280 | VTDLLASFTSLSLQM +++++++++++ | HHHHHHCCCCCCCCC | c1b-4 | multi | 0.263 | 0.384 | 0.293 | DISO | 0.0 | |
| 18 | cand_D | Q6NXT1 | 280 | VTDLLASFTSLSLQMQS +++++++++++ | HHHHHHCCCCCCCCCCC | c1c-AT-4 | multi | 0.327 | 0.441 | 0.305 | DISO | 0.0 | |
| 19 | cand_D | Q6NXT1 | 281 | TDLLASFTSLSLQM +++++++++++ | HHHHHCCCCCCCCC | c2-AT-4 | multi | 0.281 | 0.399 | 0.289 | DISO | 0.0 | |
| 20 | fp_D | Q6NXT1 | 286 | SFTSLSLQMQSMEK +++++++ | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.572 | 0.663 | 0.334 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment