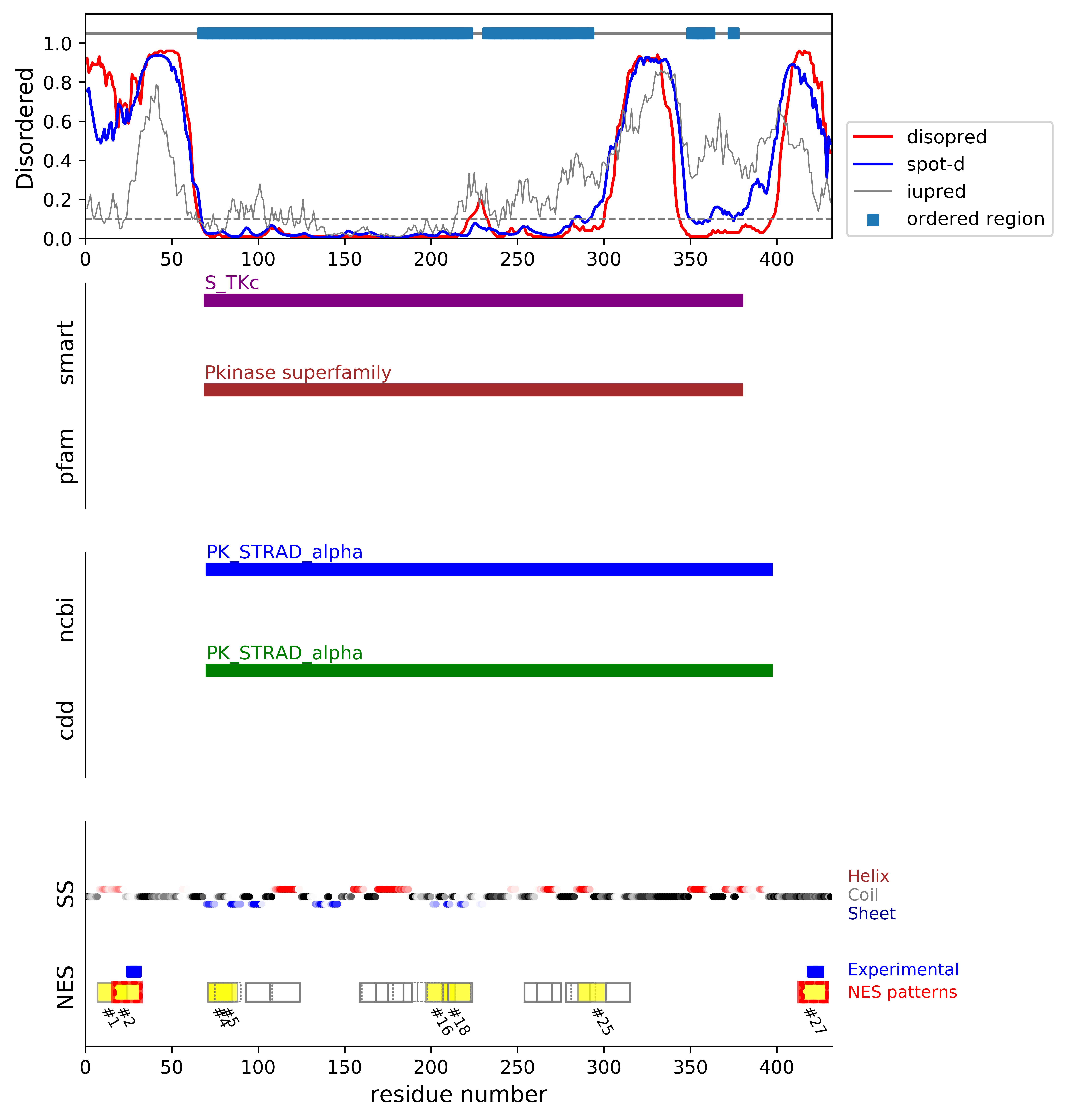

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q7RTN6 | 7 | KPERIRRWVSEKFIVEG | CHHHHHHHHHHHHHHHC | c1c-4 | uniq | 0.778 | 0.567 | 0.121 | DISO | 0.0 | |
| 2 | cand_D | Q7RTN6 | 16 | SEKFIVEGLRDLELFG * * | HHHHHHHCCCCCCCCC | c1a-4 | multi-selected | 0.711 | 0.646 | 0.209 | DISO | 0.0 | |
| 3 | cand_D | Q7RTN6 | 17 | EKFIVEGLRDLELFG * * | HHHHHHCCCCCCCCC | c1b-4 | multi | 0.706 | 0.656 | 0.207 | DISO | 0.0 | |
| 4 | fp_D | Q7RTN6 | 71 | LLTVIGKGFEDLMT | EEEEECCCCCCCCE | c3-4 | multi-selected | 0.013 | 0.026 | 0.074 | boundary | boundary|PK_STRAD_alpha; | 0.286 |
| 5 | fp_D | Q7RTN6 | 71 | LLTVIGKGFEDLMTVNL | EEEEECCCCCCCCEEEE | c1c-5 | multi-selected | 0.012 | 0.023 | 0.093 | boundary | boundary|PK_STRAD_alpha; | 0.125 |
| 6 | fp_D | Q7RTN6 | 75 | IGKGFEDLMTVNLAR | ECCCCCCCCEEEEEE | c1b-4 | multi | 0.012 | 0.021 | 0.1 | boundary | boundary|PK_STRAD_alpha; | 0.286 |
| 7 | fp_beta_O | Q7RTN6 | 93 | TGEYVTVRRINLEA | CCCEEEEEEECCCC | c2-4 | uniq | 0.015 | 0.032 | 0.159 | ORD | MID|PK_STRAD_alpha; | 1.0 |
| 8 | fp_O | Q7RTN6 | 107 | CSNEMVTFLQGELHVSK | CCHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.029 | 0.029 | 0.103 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 9 | fp_O | Q7RTN6 | 108 | SNEMVTFLQGELHVSK | CHHHHHHHHHHHHHHH | c1d-4 | multi | 0.028 | 0.027 | 0.104 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 10 | fp_O | Q7RTN6 | 159 | ICTHFMDGMNELAIAY | HHHCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.01 | 0.019 | 0.029 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 11 | fp_O | Q7RTN6 | 160 | CTHFMDGMNELAIAY | HHCCCCCCCHHHHHH | c1b-4 | multi | 0.01 | 0.019 | 0.028 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 12 | fp_O | Q7RTN6 | 168 | NELAIAYILQGVLKAL | CHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.007 | 0.008 | 0.015 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 13 | fp_O | Q7RTN6 | 175 | ILQGVLKALDYIHH | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.006 | 0.007 | 0.02 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 14 | fp_O | Q7RTN6 | 178 | GVLKALDYIHHMGY | HHHHHHHHHHCCCC | c3-AT | multi | 0.006 | 0.009 | 0.03 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 15 | fp_O | Q7RTN6 | 192 | VHRSVKASHILISV | CCCCCCCCCEEECC | c2-AT-4 | multi | 0.01 | 0.018 | 0.049 | ORD | MID|PK_STRAD_alpha; | 0.143 |
| 16 | fp_D | Q7RTN6 | 197 | KASHILISVDGKVYLSG | CCCCEEECCCCCEEECC | c1c-4 | multi-selected | 0.01 | 0.021 | 0.051 | boundary | MID|PK_STRAD_alpha; | 0.375 |
| 17 | fp_D | Q7RTN6 | 198 | ASHILISVDGKVYLSG | CCCEEECCCCCEEECC | c1d-4 | multi | 0.01 | 0.021 | 0.048 | boundary | MID|PK_STRAD_alpha; | 0.286 |
| 18 | fp_D | Q7RTN6 | 207 | GKVYLSGLRSNLSMIS | CCEEECCCCEEEEECC | c1aR-5 | multi-selected | 0.028 | 0.021 | 0.123 | boundary | MID|PK_STRAD_alpha; | 0.429 |
| 19 | fp_D | Q7RTN6 | 207 | GKVYLSGLRSNLSMIS | CCEEECCCCEEEEECC | c1d-4 | multi | 0.028 | 0.021 | 0.123 | boundary | MID|PK_STRAD_alpha; | 0.429 |
| 20 | fp_beta_D | Q7RTN6 | 210 | YLSGLRSNLSMISH | EECCCCEEEEECCC | c3-4 | multi-selected | 0.038 | 0.02 | 0.144 | boundary | MID|PK_STRAD_alpha; | 0.571 |
| 21 | fp_O | Q7RTN6 | 254 | QGYDAKSDIYSVGITA | CCCCCCCHHHHHHHHH | c1a-AT-4 | uniq | 0.016 | 0.033 | 0.201 | ORD | MID|PK_STRAD_alpha; | 0.0 |
| 22 | fp_D | Q7RTN6 | 261 | DIYSVGITACELAN | HHHHHHHHHHHHHC | c3-AT | uniq | 0.01 | 0.021 | 0.176 | boundary | MID|PK_STRAD_alpha; | 0.0 |
| 23 | fp_D | Q7RTN6 | 278 | PFKDMPATQMLLEK | CCCCCCHHHHHHHH | c2-AT-5 | multi-selected | 0.05 | 0.088 | 0.352 | boundary | MID|PK_STRAD_alpha; | 0.0 |
| 24 | fp_D | Q7RTN6 | 281 | DMPATQMLLEKLNG | CCCHHHHHHHHHCC | c3-AT | multi | 0.053 | 0.1 | 0.355 | boundary | MID|PK_STRAD_alpha; | 0.0 |
| 25 | fp_D | Q7RTN6 | 285 | TQMLLEKLNGTVPCLL | HHHHHHHHCCCCCCCC | c1aR-5 | multi-selected | 0.053 | 0.132 | 0.332 | boundary | MID|PK_STRAD_alpha; | 0.0 |
| 26 | fp_D | Q7RTN6 | 301 | DTSTIPAEELTMSP | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.472 | 0.516 | 0.484 | boundary | MID|PK_STRAD_alpha; | 0.0 |
| 27 | cand_D | Q7RTN6 | 413 | GIFGLVTNLEELEVDD * * | CCCCCCCCCCCCCCCC | c1a-5 | multi-selected | 0.85 | 0.705 | 0.323 | DISO | boundary|PK_STRAD_alpha; | 0.0 |
| 28 | cand_D | Q7RTN6 | 414 | IFGLVTNLEELEVDD * * | CCCCCCCCCCCCCCC | c1b-5 | multi | 0.843 | 0.694 | 0.313 | DISO | boundary|PK_STRAD_alpha; | 0.0 |
| 29 | cand_D | Q7RTN6 | 415 | FGLVTNLEELEVDD * * | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.835 | 0.687 | 0.299 | DISO | boundary|PK_STRAD_alpha; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment