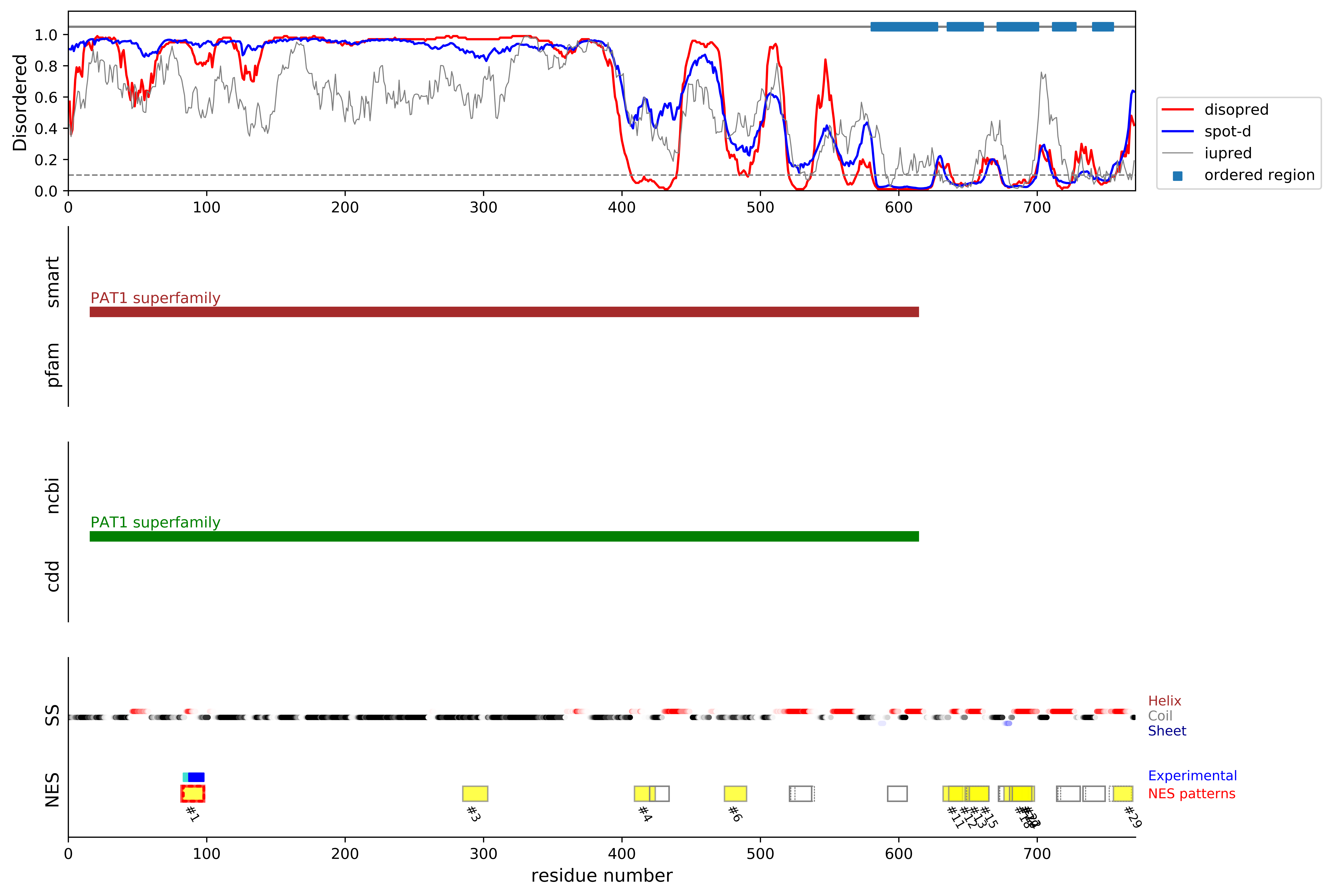

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | Q86TB9 | 82 | HEENLAERLSKMVIEN ++++*++*+* | CCCHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.883 | 0.945 | 0.572 | DISO | MID|PAT1 superfamily; | 0.0 |
| 2 | cand_D | Q86TB9 | 83 | EENLAERLSKMVIEN ++++*++*+* | CCHHHHHHHHCCCCC | c1b-AT-4 | multi | 0.878 | 0.944 | 0.563 | DISO | MID|PAT1 superfamily; | 0.0 |
| 3 | fp_D | Q86TB9 | 285 | VPGFVGSPLAAMNPKLLQ | CCCCCCCCCCCCCCCCCC | c4-4 | uniq | 0.972 | 0.872 | 0.592 | DISO | MID|PAT1 superfamily; | 0.0 |
| 4 | fp_D | Q86TB9 | 409 | EKDWVSKIQMMQLQS | HHHHHHHCCCCCCCC | c1b-4 | uniq | 0.063 | 0.517 | 0.458 | DISO | MID|PAT1 superfamily; | 0.0 |
| 5 | fp_D | Q86TB9 | 420 | QLQSTDPYLDDFYY | CCCCCCCCCCHHHH | c3-AT | uniq | 0.035 | 0.483 | 0.356 | DISO | MID|PAT1 superfamily; | 0.0 |
| 6 | fp_D | Q86TB9 | 474 | KPVQFEGSLGKLTVSS | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.211 | 0.317 | 0.402 | DISO | MID|PAT1 superfamily; | 0.0 |
| 7 | fp_D | Q86TB9 | 521 | KTLVIIEKTYSLLLDV | HHHHHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.034 | 0.163 | 0.153 | DISO | MID|PAT1 superfamily; | 0.0 |
| 8 | fp_D | Q86TB9 | 522 | TLVIIEKTYSLLLDV | HHHHHHHHHHHHHHH | c1b-AT-5 | multi | 0.031 | 0.161 | 0.14 | DISO | MID|PAT1 superfamily; | 0.0 |
| 9 | fp_D | Q86TB9 | 525 | IIEKTYSLLLDVED | HHHHHHHHHHHHHH | c3-AT | multi | 0.046 | 0.166 | 0.118 | DISO | MID|PAT1 superfamily; | 0.0 |
| 10 | fp_D | Q86TB9 | 592 | KGKRMVARILPFLS | CHHHHHHHHCCCCC | c2-AT-4 | uniq | 0.011 | 0.025 | 0.115 | boundary | boundary|PAT1 superfamily; | 0.0 |
| 11 | fp_D | Q86TB9 | 632 | VLPCLLSPFSLLLY | CCCCCHHHHHHHHH | c3-4 | multi-selected | 0.096 | 0.062 | 0.043 | boundary | boundary|PAT1 superfamily; | 0.0 |
| 12 | fp_D | Q86TB9 | 636 | LLSPFSLLLYHLPS | CHHHHHHHHHCCCH | c3-4 | multi-selected | 0.062 | 0.041 | 0.035 | boundary | boundary|PAT1 superfamily; | 0.0 |
| 13 | fp_D | Q86TB9 | 648 | PSVSITSLLRQLMNLPQ | CHHHHHHHHHHHHHCCC | c1c-4 | multi-selected | 0.094 | 0.062 | 0.158 | boundary | boundary|PAT1 superfamily; | 0.0 |
| 14 | fp_D | Q86TB9 | 649 | SVSITSLLRQLMNLPQ | HHHHHHHHHHHHHCCC | c1d-AT-5 | multi | 0.097 | 0.063 | 0.165 | boundary | boundary|PAT1 superfamily; | 0.0 |
| 15 | fp_D | Q86TB9 | 651 | SITSLLRQLMNLPQ | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.106 | 0.066 | 0.184 | boundary | 0.0 | |
| 16 | fp_beta_D | Q86TB9 | 672 | SNPHLTAVLQNKFGLSL | CCCCEEEEECCHHHHHH | c1c-4 | multi-selected | 0.052 | 0.049 | 0.138 | boundary | 0.625 | |
| 17 | fp_beta_D | Q86TB9 | 673 | NPHLTAVLQNKFGLSL | CCCEEEEECCHHHHHH | c1d-AT-4 | multi | 0.046 | 0.042 | 0.124 | boundary | 0.571 | |
| 18 | fp_D | Q86TB9 | 676 | LTAVLQNKFGLSLLLIL | EEEEECCHHHHHHHHHH | c1c-4 | multi-selected | 0.042 | 0.028 | 0.065 | boundary | 0.125 | |
| 19 | fp_D | Q86TB9 | 680 | LQNKFGLSLLLILL | ECCHHHHHHHHHHH | c2-4 | multi-selected | 0.046 | 0.027 | 0.031 | boundary | 0.0 | |
| 20 | fp_D | Q86TB9 | 680 | LQNKFGLSLLLILLSR | ECCHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.048 | 0.028 | 0.038 | boundary | 0.0 | |
| 21 | fp_D | Q86TB9 | 682 | NKFGLSLLLILLSRGE | CHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.055 | 0.031 | 0.052 | boundary | 0.0 | |
| 22 | fp_D | Q86TB9 | 682 | NKFGLSLLLILLSR | CHHHHHHHHHHHHH | c2-4 | multi-selected | 0.051 | 0.028 | 0.037 | boundary | 0.0 | |
| 23 | fp_D | Q86TB9 | 714 | WTEVMFMATRELLRIPQ | HHHHHHHHHHHHHHCCC | c1c-AT-4 | multi-selected | 0.073 | 0.062 | 0.176 | boundary | 0.0 | |
| 24 | fp_D | Q86TB9 | 715 | TEVMFMATRELLRIPQ | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.074 | 0.061 | 0.158 | boundary | 0.0 | |
| 25 | fp_D | Q86TB9 | 717 | VMFMATRELLRIPQ | HHHHHHHHHHHCCC | c3-AT | multi | 0.081 | 0.06 | 0.135 | boundary | 0.0 | |
| 26 | fp_D | Q86TB9 | 733 | LAKPISIPTNLVSLFS | CCCCCCCCCCHHHHHH | c1a-AT-4 | multi-selected | 0.152 | 0.093 | 0.095 | boundary | 0.0 | |
| 27 | fp_D | Q86TB9 | 735 | KPISIPTNLVSLFS | CCCCCCCCHHHHHH | c2-AT-4 | multi | 0.138 | 0.089 | 0.091 | boundary | 0.0 | |
| 28 | fp_D | Q86TB9 | 752 | DRQKLNLLETKLQLVQ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.177 | 0.225 | 0.116 | boundary | 0.0 | |
| 29 | fp_D | Q86TB9 | 755 | KLNLLETKLQLVQG | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.221 | 0.279 | 0.119 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment