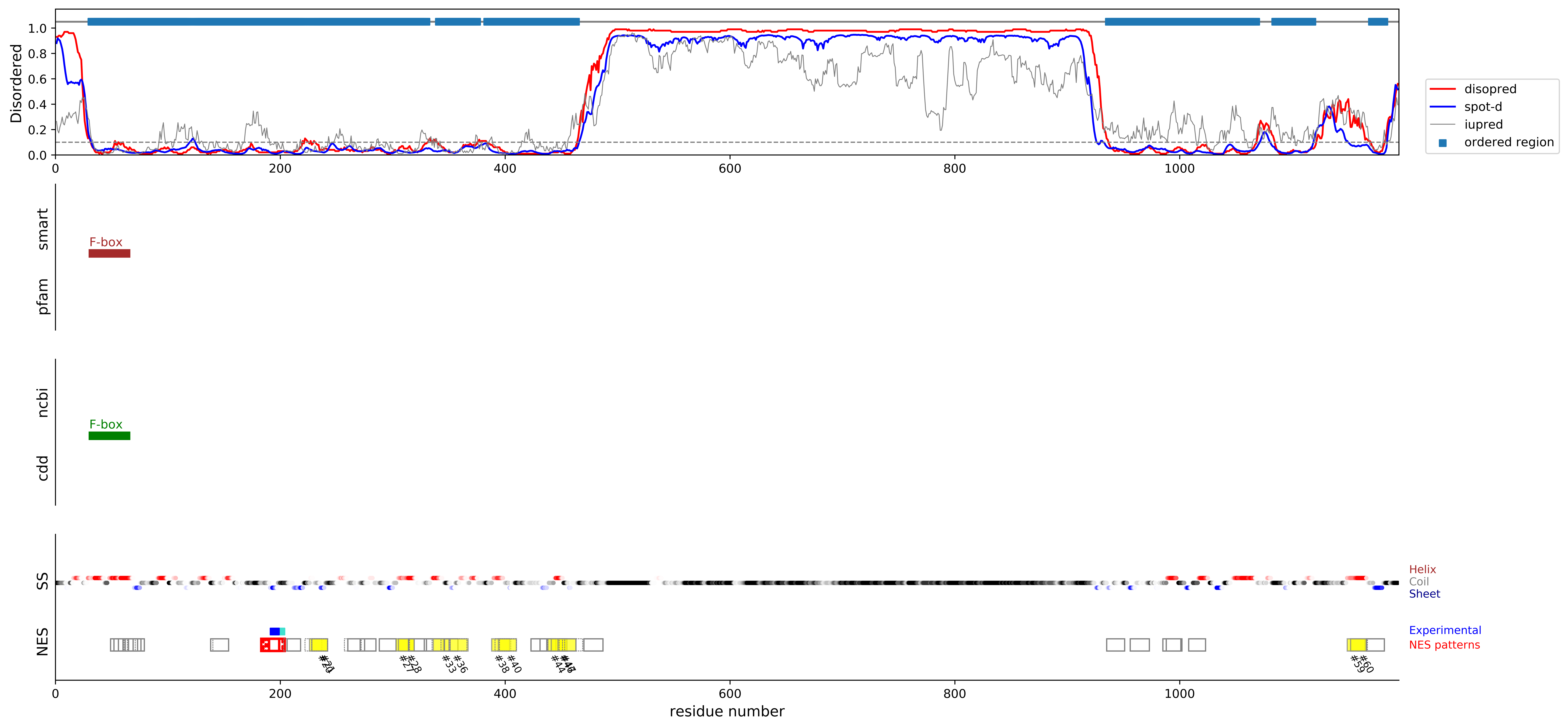

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_O | Q8BMI0 | 49 | DIMCMECLSRKLKEAV | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.073 | 0.037 | 0.034 | ORD | 0.0 | |
| 2 | fp_O | Q8BMI0 | 52 | CMECLSRKLKEAVTLYL | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.072 | 0.031 | 0.035 | ORD | 0.0 | |
| 3 | fp_O | Q8BMI0 | 56 | LSRKLKEAVTLYLRV | HHHHHHHHHHHHHHE | c1b-AT-4 | multi | 0.065 | 0.026 | 0.03 | ORD | 0.0 | |
| 4 | fp_O | Q8BMI0 | 56 | LSRKLKEAVTLYLRVVR | HHHHHHHHHHHHHHEEE | c1c-4 | multi-selected | 0.06 | 0.024 | 0.029 | ORD | 0.0 | |
| 5 | fp_O | Q8BMI0 | 60 | LKEAVTLYLRVVRVVD | HHHHHHHHHHEEEEEE | c1a-4 | multi-selected | 0.044 | 0.019 | 0.022 | ORD | 0.143 | |
| 6 | fp_O | Q8BMI0 | 61 | KEAVTLYLRVVRVVD | HHHHHHHHHEEEEEE | c1b-AT-4 | multi | 0.041 | 0.019 | 0.022 | ORD | 0.286 | |
| 7 | fp_beta_O | Q8BMI0 | 62 | EAVTLYLRVVRVVDLCA | HHHHHHHHEEEEEECCC | c1c-4 | multi-selected | 0.034 | 0.018 | 0.022 | ORD | 0.5 | |
| 8 | fp_O | Q8BMI0 | 62 | EAVTLYLRVVRVVD | HHHHHHHHEEEEEE | c2-4 | multi-selected | 0.039 | 0.018 | 0.022 | ORD | 0.286 | |
| 9 | fp_beta_O | Q8BMI0 | 64 | VTLYLRVVRVVDLCA | HHHHHHEEEEEECCC | c1b-4 | multi | 0.03 | 0.017 | 0.02 | ORD | 0.714 | |
| 10 | fp_O | Q8BMI0 | 138 | CPNLVGVETSHLELVE | CCCCCCCCCCCCHHHH | c1a-AT-4 | multi-selected | 0.029 | 0.032 | 0.072 | ORD | 0.0 | |
| 11 | fp_O | Q8BMI0 | 140 | NLVGVETSHLELVE | CCCCCCCCCCHHHH | c2-AT-5 | multi | 0.031 | 0.032 | 0.067 | ORD | 0.0 | |
| 12 | cand_O | Q8BMI0 | 183 | LKIPIGAKIQTLHLVG *+*++ | CCCCCCCCEEEEEEEC | c1a-4 | multi-selected | 0.021 | 0.025 | 0.128 | ORD | 0.429 | |
| 13 | cand_beta_O | Q8BMI0 | 185 | IPIGAKIQTLHLVG *+*++ | CCCCCCEEEEEEEC | c2-AT-4 | multi | 0.021 | 0.023 | 0.119 | ORD | 0.571 | |
| 14 | cand_beta_O | Q8BMI0 | 187 | IGAKIQTLHLVGVNV *+*+++++ | CCCCEEEEEEECCCC | c1b-4 | multi | 0.023 | 0.019 | 0.096 | ORD | 1.0 | |
| 15 | cand_beta_O | Q8BMI0 | 189 | AKIQTLHLVGVNVPE *+*+++++ | CCEEEEEEECCCCCC | c1b-AT-4 | multi | 0.024 | 0.018 | 0.083 | ORD | 0.714 | |
| 16 | cand_beta_O | Q8BMI0 | 190 | KIQTLHLVGVNVPE *+*+++++ | CEEEEEEECCCCCC | c2-5 | multi-selected | 0.024 | 0.017 | 0.08 | ORD | 0.714 | |
| 17 | fp_D | Q8BMI0 | 203 | EIPCIPMLRHLYMKW | CCCCCCCCCEEEEEE | c1b-5 | multi | 0.026 | 0.017 | 0.04 | boundary | 0.286 | |
| 18 | fp_beta_D | Q8BMI0 | 206 | CIPMLRHLYMKW | CCCCCCEEEEEE | c2-rev | multi-selected | 0.027 | 0.014 | 0.041 | boundary | 0.5 | |
| 19 | fp_D | Q8BMI0 | 222 | KPQPFKDFLCISLRT | CCCCCHHHCCCCCCE | c1b-4 | multi | 0.091 | 0.037 | 0.033 | boundary | 0.0 | |
| 20 | fp_D | Q8BMI0 | 226 | FKDFLCISLRTFVMRN | CHHHCCCCCCEEEECC | c1a-4 | multi-selected | 0.071 | 0.029 | 0.043 | boundary | 0.143 | |
| 21 | fp_D | Q8BMI0 | 228 | DFLCISLRTFVMRN | HHCCCCCCEEEECC | c2-5 | multi-selected | 0.067 | 0.027 | 0.044 | __ | 0.286 | |
| 22 | fp_D | Q8BMI0 | 228 | DFLCISLRTFVMRN | HHCCCCCCEEEECC | c3-AT | multi | 0.067 | 0.027 | 0.044 | __ | 0.286 | |
| 23 | fp_O | Q8BMI0 | 257 | GLASARNLEHLEMVR | HHHCCCCCCCEECCC | c1b-AT-5 | multi | 0.038 | 0.047 | 0.078 | ORD | 0.143 | |
| 24 | fp_O | Q8BMI0 | 260 | SARNLEHLEMVRVPF | CCCCCCCEECCCCCC | c1b-4 | multi-selected | 0.033 | 0.044 | 0.07 | ORD | 0.286 | |
| 25 | fp_O | Q8BMI0 | 271 | RVPFLGGLIQHVVE | CCCCCCCCHHHHCC | c3-4 | uniq | 0.039 | 0.035 | 0.051 | ORD | 0.0 | |
| 26 | fp_O | Q8BMI0 | 288 | RSGGFRNLHTIVLGA | CCCCCCCCEEEEECC | c1b-4 | uniq | 0.025 | 0.028 | 0.065 | ORD | 0.429 | |
| 27 | fp_D | Q8BMI0 | 303 | CKNALEVDLGYLIITA | CHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.051 | 0.025 | 0.021 | boundary | 0.0 | |
| 28 | fp_D | Q8BMI0 | 305 | NALEVDLGYLIITA | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.054 | 0.024 | 0.02 | boundary | 0.0 | |
| 29 | fp_D | Q8BMI0 | 314 | LIITAARRLHEVRIQP | HHHHHHHHCCCCCCCC | c1a-AT-5 | multi-selected | 0.047 | 0.027 | 0.084 | boundary | 0.0 | |
| 30 | fp_D | Q8BMI0 | 315 | IITAARRLHEVRIQP | HHHHHHHCCCCCCCC | c1b-AT-5 | multi | 0.046 | 0.027 | 0.087 | boundary | 0.0 | |
| 31 | fp_D | Q8BMI0 | 328 | QPSLTKDGVFSALKMAE | CCCCCCHHHHHHHHHHH | c1c-AT-4 | uniq | 0.097 | 0.039 | 0.107 | boundary | 0.0 | |
| 32 | fp_D | Q8BMI0 | 335 | GVFSALKMAELEFPQ | HHHHHHHHHHCCCCC | c1b-AT-5 | multi | 0.077 | 0.032 | 0.066 | boundary | 0.0 | |
| 33 | fp_D | Q8BMI0 | 336 | VFSALKMAELEFPQ | HHHHHHHHHCCCCC | c2-5 | multi-selected | 0.074 | 0.032 | 0.066 | __ | 0.0 | |
| 34 | fp_D | Q8BMI0 | 343 | AELEFPQFETLHLGY | HHCCCCCCCEEEECC | c1b-4 | multi | 0.033 | 0.027 | 0.031 | boundary | 0.286 | |
| 35 | fp_beta_D | Q8BMI0 | 346 | EFPQFETLHLGY | CCCCCCEEEECC | c2-rev | multi-selected | 0.026 | 0.025 | 0.027 | boundary | 0.5 | |
| 36 | fp_D | Q8BMI0 | 351 | ETLHLGYVDEFLLQSR | CEEEECCCCHHHHHHC | c1aR-4 | multi-selected | 0.041 | 0.037 | 0.028 | boundary | 0.143 | |
| 37 | fp_D | Q8BMI0 | 351 | ETLHLGYVDEFLLQS | CEEEECCCCHHHHHH | c1b-4 | multi | 0.039 | 0.035 | 0.028 | boundary | 0.143 | |
| 38 | fp_D | Q8BMI0 | 388 | TDIGMKAVNEVFSCIK | CHHHHHHHHHHCCCCC | c1aR-5 | multi-selected | 0.045 | 0.018 | 0.042 | boundary | 0.0 | |
| 39 | fp_D | Q8BMI0 | 391 | GMKAVNEVFSCIKY | HHHHHHHHCCCCCE | c3-4 | multi | 0.037 | 0.015 | 0.026 | boundary | 0.0 | |
| 40 | fp_D | Q8BMI0 | 394 | AVNEVFSCIKYLAIYN | HHHHHCCCCCEECCCC | c1a-5 | multi-selected | 0.024 | 0.012 | 0.017 | boundary | 0.143 | |
| 41 | fp_D | Q8BMI0 | 395 | VNEVFSCIKYLAIYN | HHHHCCCCCEECCCC | c1b-4 | multi | 0.022 | 0.012 | 0.017 | boundary | 0.286 | |
| 42 | fp_O | Q8BMI0 | 423 | HSRWMRLVDINLVR | CCCCCCCCCCEEEE | c2-4 | uniq | 0.01 | 0.018 | 0.071 | ORD | 0.0 | |
| 43 | fp_beta_O | Q8BMI0 | 431 | DINLVRCHALKLDS | CCEEEEECCCCCCH | c3-AT | uniq | 0.014 | 0.014 | 0.055 | ORD | 0.571 | |
| 44 | fp_D | Q8BMI0 | 438 | HALKLDSFGQFVELLP | CCCCCCHHHHHHHHCC | c1aR-5 | multi-selected | 0.019 | 0.025 | 0.059 | boundary | 0.0 | |
| 45 | fp_D | Q8BMI0 | 438 | HALKLDSFGQFVELLP | CCCCCCHHHHHHHHCC | c1d-4 | multi | 0.019 | 0.025 | 0.059 | boundary | 0.0 | |
| 46 | fp_D | Q8BMI0 | 441 | KLDSFGQFVELLPS | CCCHHHHHHHHCCC | c3-4 | multi-selected | 0.02 | 0.028 | 0.062 | boundary | 0.0 | |
| 47 | fp_D | Q8BMI0 | 447 | QFVELLPSLEFISLDQ | HHHHHCCCCCEECCCC | c1a-5 | multi-selected | 0.021 | 0.034 | 0.125 | boundary | 0.143 | |
| 48 | fp_D | Q8BMI0 | 448 | FVELLPSLEFISLDQ | HHHHCCCCCEECCCC | c1b-5 | multi | 0.021 | 0.034 | 0.13 | boundary | 0.286 | |
| 49 | fp_D | Q8BMI0 | 451 | LLPSLEFISLDQ | HCCCCCEECCCC | c2-rev | multi | 0.019 | 0.036 | 0.15 | boundary | 0.333 | |
| 50 | fp_D | Q8BMI0 | 453 | PSLEFISLDQMFREPP | CCCCEECCCCCCCCCC | c1aR-4 | multi | 0.085 | 0.06 | 0.203 | boundary | 0.286 | |
| 51 | fp_D | Q8BMI0 | 470 | GCARVGLSAGTGIGVSS | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.632 | 0.423 | 0.379 | boundary | 0.0 | |
| 52 | fp_D | Q8BMI0 | 935 | GVTMTNCGITDLVLKD | CEEEEECCCCCCCCCC | c1a-AT-5 | uniq | 0.053 | 0.051 | 0.187 | boundary | 0.286 | |
| 53 | fp_O | Q8BMI0 | 956 | FIHATRCRVLKHLKVEN | EEEEECCCCCCCCCCCC | c1c-AT-5 | uniq | 0.025 | 0.038 | 0.168 | ORD | 0.125 | |
| 54 | fp_O | Q8BMI0 | 985 | KKLNMDQVLDQILRMP | CCCCHHHHHHHHHCCC | c1aR-5 | multi-selected | 0.033 | 0.03 | 0.228 | ORD | 0.0 | |
| 55 | fp_O | Q8BMI0 | 985 | KKLNMDQVLDQILRMPP | CCCCHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.034 | 0.031 | 0.227 | ORD | 0.0 | |
| 56 | fp_O | Q8BMI0 | 985 | KKLNMDQVLDQILRMPP | CCCCHHHHHHHHHCCCC | c1cR-4 | multi | 0.034 | 0.031 | 0.227 | ORD | 0.0 | |
| 57 | fp_O | Q8BMI0 | 988 | NMDQVLDQILRMPP | CHHHHHHHHHCCCC | c3-4 | multi-selected | 0.039 | 0.028 | 0.246 | ORD | 0.0 | |
| 58 | fp_O | Q8BMI0 | 1008 | YLRPMQQVDTLTLEQ | EEECCCCCCHHHHHH | c1b-5 | uniq | 0.043 | 0.022 | 0.214 | ORD | 0.0 | |
| 59 | fp_D | Q8BMI0 | 1149 | ADICMETIGEEISEMR | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.271 | 0.078 | 0.325 | boundary | 0.0 | |
| 60 | fp_D | Q8BMI0 | 1152 | CMETIGEEISEMRQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.234 | 0.073 | 0.313 | boundary | 0.0 | |
| 61 | fp_beta_D | Q8BMI0 | 1166 | MKRGIFQRVVAIFIHY | HCCCCCCEEEEEEEEC | c1a-4 | multi-selected | 0.061 | 0.03 | 0.086 | boundary | 0.571 | |
| 62 | fp_beta_D | Q8BMI0 | 1167 | KRGIFQRVVAIFIHY | CCCCCCEEEEEEEEC | c1b-4 | multi | 0.057 | 0.027 | 0.077 | boundary | 0.714 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment