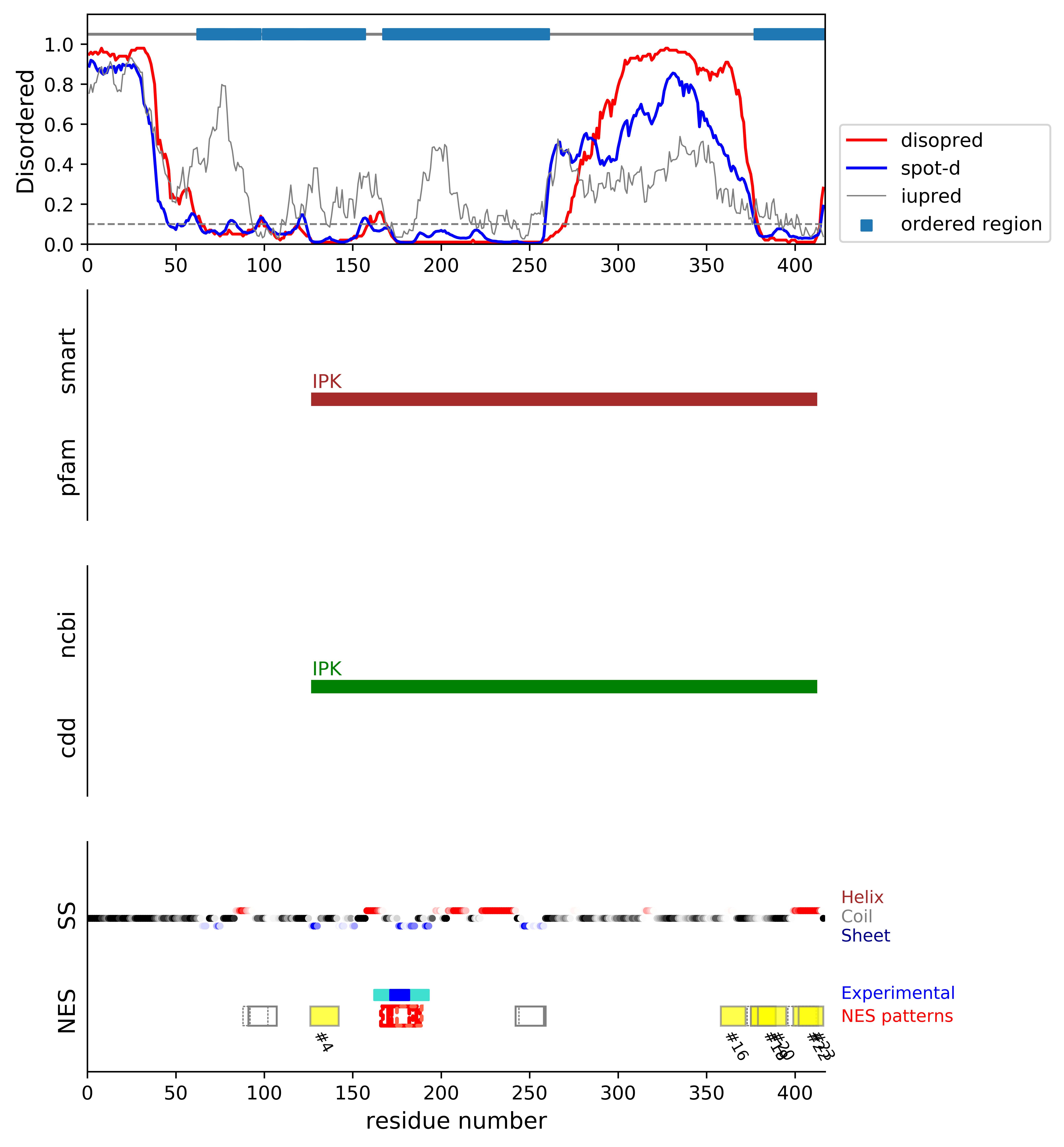

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q8NFU5 | 88 | EFYNMVYAADCFDG | HHHHHHCCCCCCCC | c3-AT | multi | 0.081 | 0.085 | 0.13 | boundary | 0.0 | |
| 2 | fp_D | Q8NFU5 | 91 | NMVYAADCFDGVLLEL | HHHCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.082 | 0.086 | 0.078 | boundary | boundary|IPK; | 0.0 |
| 3 | fp_D | Q8NFU5 | 92 | MVYAADCFDGVLLEL | HHCCCCCCCCCCCCC | c1b-AT-5 | multi | 0.083 | 0.088 | 0.067 | boundary | boundary|IPK; | 0.0 |
| 4 | fp_D | Q8NFU5 | 126 | LYLKLEDVTHKFNKPC | EEEEECCCCCCCCCCC | c1aR-4 | uniq | 0.011 | 0.018 | 0.224 | boundary | boundary|IPK; | 0.143 |
| 5 | cand_D | Q8NFU5 | 166 | SKYPLMEEIGFLVLGM ++++++++*++*+*++ | HHCCCCCCCCEEEEEE | c1a-4 | multi | 0.051 | 0.041 | 0.082 | boundary | MID|IPK; | 0.143 |
| 6 | cand_D | Q8NFU5 | 167 | KYPLMEEIGFLVLGM +++++++*++*+*++ | HCCCCCCCCEEEEEE | c1b-4 | multi | 0.043 | 0.039 | 0.072 | boundary | MID|IPK; | 0.286 |
| 7 | cand_beta_D | Q8NFU5 | 170 | LMEEIGFLVLGMRV ++++*++*+*++++ | CCCCCCEEEEEEEE | c2-5 | multi | 0.023 | 0.026 | 0.058 | boundary | MID|IPK; | 0.571 |
| 8 | cand_beta_D | Q8NFU5 | 170 | LMEEIGFLVLGMRVYH ++++*++*+*++++++ | CCCCCCEEEEEEEEEE | c1a-5 | multi-selected | 0.021 | 0.025 | 0.062 | boundary | MID|IPK; | 0.714 |
| 9 | cand_beta_D | Q8NFU5 | 170 | LMEEIGFLVLGMRVYH ++++*++*+*++++++ | CCCCCCEEEEEEEEEE | c1d-5 | multi | 0.021 | 0.025 | 0.062 | boundary | MID|IPK; | 0.714 |
| 10 | cand_beta_D | Q8NFU5 | 172 | EEIGFLVLGMRVYH ++*++*+*++++++ | CCCCEEEEEEEEEE | c2-4 | multi | 0.014 | 0.017 | 0.061 | boundary | MID|IPK; | 0.857 |
| 11 | cand_beta_D | Q8NFU5 | 172 | EEIGFLVLGMRVYHVH ++*++*+*++++++++ | CCCCEEEEEEEEEECC | c1aR-5 | multi | 0.014 | 0.019 | 0.075 | boundary | MID|IPK; | 1.0 |
| 12 | cand_beta_D | Q8NFU5 | 175 | GFLVLGMRVYHVHS ++*+*+++++++++ | CEEEEEEEEEECCC | c3-4 | multi | 0.01 | 0.017 | 0.086 | boundary | MID|IPK; | 1.0 |
| 13 | fp_beta_D | Q8NFU5 | 242 | NQKQLNFYASSLLFVYE | HCCCEEEEEEEEEEEEE | c1c-AT-4 | multi-selected | 0.011 | 0.009 | 0.105 | boundary | MID|IPK; | 1.0 |
| 14 | fp_beta_D | Q8NFU5 | 242 | NQKQLNFYASSLLFVY | HCCCEEEEEEEEEEEE | c1a-AT-4 | multi-selected | 0.01 | 0.007 | 0.103 | boundary | MID|IPK; | 1.0 |
| 15 | fp_beta_D | Q8NFU5 | 244 | KQLNFYASSLLFVY | CCEEEEEEEEEEEE | c2-AT-4 | multi | 0.01 | 0.006 | 0.104 | boundary | MID|IPK; | 1.0 |
| 16 | fp_D | Q8NFU5 | 358 | VLSQLEKVFYHLPT | CCCCHHHHCCCCCC | c3-4 | uniq | 0.811 | 0.392 | 0.214 | boundary | MID|IPK; | 0.0 |
| 17 | fp_D | Q8NFU5 | 373 | CQEIAEVEVRMIDFAH | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi | 0.135 | 0.094 | 0.159 | boundary | MID|IPK; | 0.0 |
| 18 | fp_D | Q8NFU5 | 375 | EIAEVEVRMIDFAH | CCCCCCCCCCCCCC | c2-5 | multi-selected | 0.098 | 0.071 | 0.158 | boundary | MID|IPK; | 0.0 |
| 19 | fp_D | Q8NFU5 | 375 | EIAEVEVRMIDFAH | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.098 | 0.071 | 0.158 | boundary | MID|IPK; | 0.0 |
| 20 | fp_D | Q8NFU5 | 379 | VEVRMIDFAHVFPSNT | CCCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.033 | 0.054 | 0.142 | boundary | boundary|IPK; | 0.0 |
| 21 | fp_D | Q8NFU5 | 396 | DEGYVYGLKHLISVLR | CCCHHHHHHHHHHHHH | c1d-4 | multi | 0.012 | 0.034 | 0.079 | boundary | boundary|IPK; | 0.0 |
| 22 | fp_D | Q8NFU5 | 399 | YVYGLKHLISVLRS | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.013 | 0.033 | 0.073 | terminal | boundary|IPK; | 0.0 |
| 23 | fp_D | Q8NFU5 | 402 | GLKHLISVLRSILD | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.035 | 0.043 | 0.066 | terminal | boundary|IPK; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment