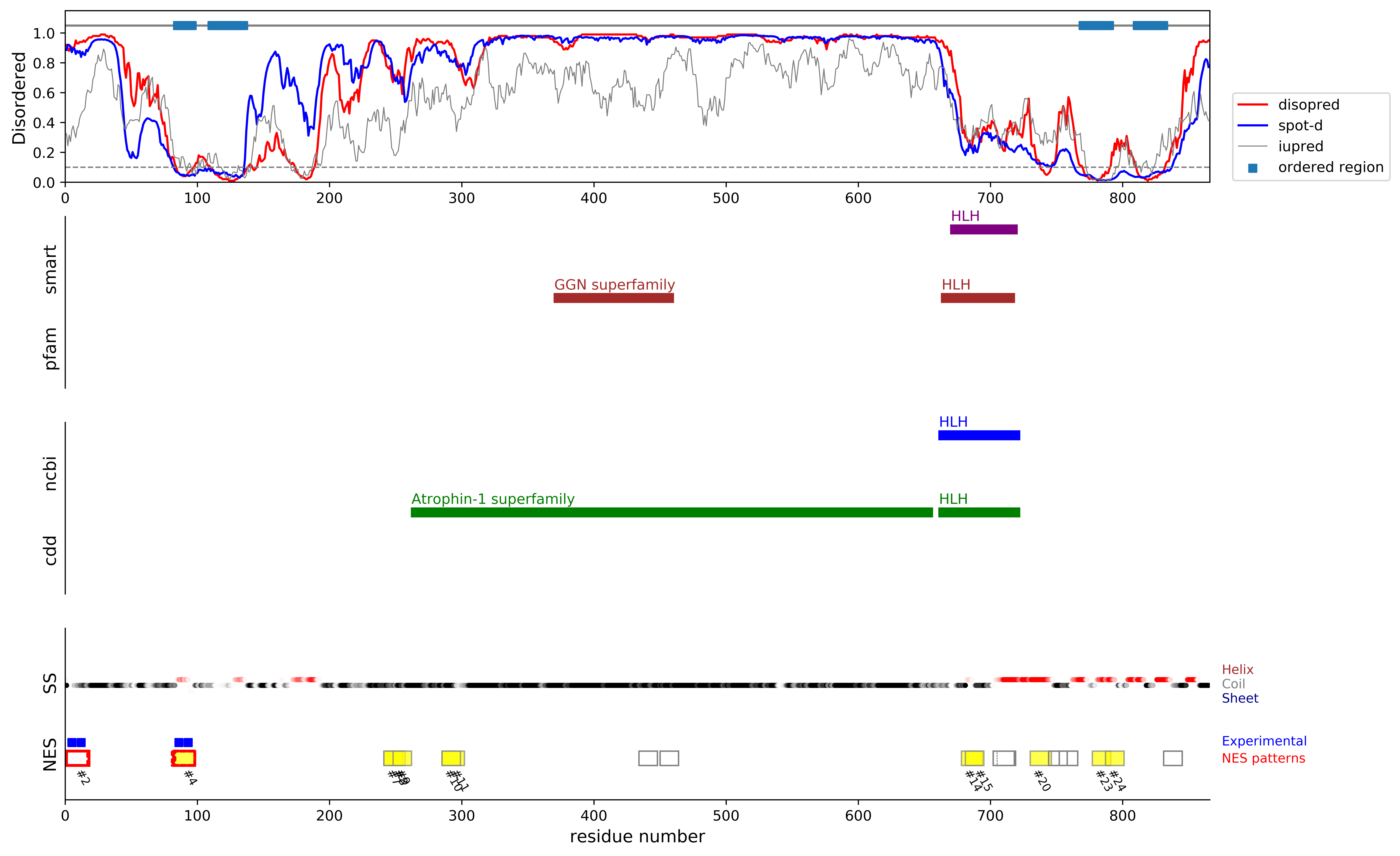

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | Q8VIP2 | 1 | MARALADLSVNLQVPR * * | CHHHHCCCCCCCCCCC | c1d-4 | multi | 0.924 | 0.881 | 0.386 | DISO | 0.0 | |
| 2 | cand_D | Q8VIP2 | 1 | XMARALADLSVNLQVPR * * | CHHHHCCCCCCCCCCCC | c1c-AT-5-Nt | multi-selected | 0.926 | 0.881 | 0.399 | DISO | 0.0 | |
| 3 | cand_D | Q8VIP2 | 81 | SIDPTLTRLFECLSLAY * * | CCCHHHHHHHHHHCEEC | c1c-AT-5 | multi | 0.079 | 0.06 | 0.14 | boundary | 0.0 | |
| 4 | cand_D | Q8VIP2 | 82 | IDPTLTRLFECLSLAY * * | CCHHHHHHHHHHCEEC | c1a-4 | multi-selected | 0.073 | 0.056 | 0.131 | boundary | 0.0 | |
| 5 | cand_D | Q8VIP2 | 82 | IDPTLTRLFECLSLAY * * | CCHHHHHHHHHHCEEC | c1d-4 | multi | 0.073 | 0.056 | 0.131 | boundary | 0.0 | |
| 6 | cand_D | Q8VIP2 | 83 | DPTLTRLFECLSLAY * * | CHHHHHHHHHHCEEC | c1b-AT-4 | multi | 0.067 | 0.053 | 0.127 | boundary | 0.0 | |
| 7 | fp_D | Q8VIP2 | 241 | QLLDLDCFLSDISD | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.744 | 0.768 | 0.313 | DISO | boundary|Atrophin-1 superfamily; | 0.0 |
| 8 | fp_D | Q8VIP2 | 241 | QLLDLDCFLSDISDTL | CCCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.729 | 0.756 | 0.321 | DISO | boundary|Atrophin-1 superfamily; | 0.0 |
| 9 | fp_D | Q8VIP2 | 248 | FLSDISDTLFTMTQ | CCCCCCCCCCCCCC | c3-4 | uniq | 0.745 | 0.655 | 0.333 | DISO | boundary|Atrophin-1 superfamily; | 0.0 |
| 10 | fp_D | Q8VIP2 | 285 | DLTPLQPSLDDFMEISD | CCCCCCCCCCCCCCCCC | c1c-5 | multi-selected | 0.866 | 0.82 | 0.51 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 11 | fp_D | Q8VIP2 | 285 | DLTPLQPSLDDFME | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.893 | 0.828 | 0.518 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 12 | fp_D | Q8VIP2 | 434 | PLFSARFPFTTVPP | CCCCCCCCCCCCCC | c3-AT | uniq | 0.966 | 0.956 | 0.538 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 13 | fp_D | Q8VIP2 | 450 | GVSTLPAPTTFVPT | CCCCCCCCCCCCCC | c3-AT | uniq | 0.982 | 0.975 | 0.778 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 14 | fp_D | Q8VIP2 | 678 | IKLGFDTLHGLVSTLS | CCCCHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.306 | 0.23 | 0.333 | DISO | boundary|Atrophin-1 superfamily; boundary|HLH; | 0.0 |
| 15 | fp_D | Q8VIP2 | 681 | GFDTLHGLVSTLSA | CHHHHHCCCCCCCC | c3-4 | multi-selected | 0.296 | 0.238 | 0.334 | DISO | boundary|Atrophin-1 superfamily; boundary|HLH; | 0.0 |
| 16 | fp_D | Q8VIP2 | 702 | KATTLQKTAEYILMLQQ | CCHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.319 | 0.258 | 0.335 | DISO | boundary|HLH; | 0.0 |
| 17 | fp_D | Q8VIP2 | 702 | KATTLQKTAEYILMLQ | CCHHHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.317 | 0.261 | 0.332 | DISO | boundary|HLH; | 0.0 |
| 18 | fp_D | Q8VIP2 | 702 | KATTLQKTAEYILMLQ | CCHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.317 | 0.261 | 0.332 | DISO | boundary|HLH; | 0.0 |
| 19 | fp_D | Q8VIP2 | 705 | TLQKTAEYILMLQQ | HHHHHHHHHHHHHH | c3-AT | multi | 0.306 | 0.248 | 0.323 | DISO | boundary|HLH; | 0.0 |
| 20 | fp_D | Q8VIP2 | 730 | LRDEIEELNAAINLCQ | HHHHHHHHHHHHHHHH | c1d-4 | uniq | 0.144 | 0.132 | 0.327 | DISO | boundary|HLH; | 0.0 |
| 21 | fp_D | Q8VIP2 | 744 | CQQQLPATGVPITH | HHHHCCCCCCCCCC | c2-AT-4 | uniq | 0.282 | 0.161 | 0.342 | boundary | boundary|HLH; | 0.0 |
| 22 | fp_D | Q8VIP2 | 752 | GVPITHQRFDQMRD | CCCCCCCCCCHHHH | c3-AT | uniq | 0.432 | 0.174 | 0.364 | boundary | boundary|HLH; | 0.0 |
| 23 | fp_D | Q8VIP2 | 777 | HNWKFWVFSILIRP | CCCHHHHHHHHHHH | c2-4 | uniq | 0.038 | 0.018 | 0.018 | boundary | 0.0 | |
| 24 | fp_D | Q8VIP2 | 787 | LIRPLFESFNGMVS | HHHHHHHHCCCCCC | c3-4 | uniq | 0.181 | 0.034 | 0.086 | boundary | 0.0 | |
| 25 | fp_D | Q8VIP2 | 831 | SLRQLSTSTSILTD | HHHHHHCCCCCCCC | c3-AT | uniq | 0.214 | 0.128 | 0.306 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment