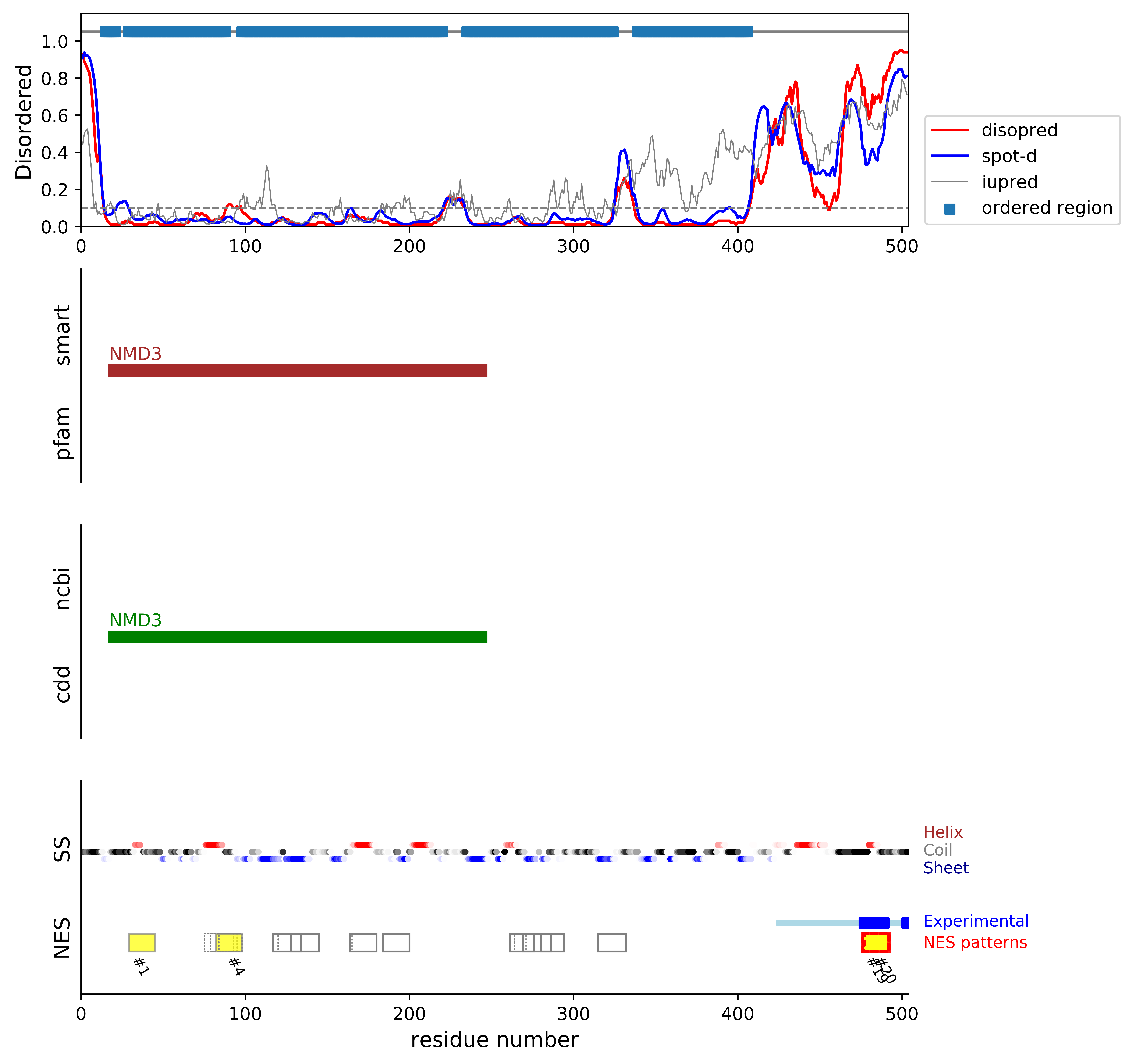

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q96D46 | 29 | ANICVACLRSKVDISQ | CCCCHHHHCCCCCCCC | c1d-4 | uniq | 0.015 | 0.054 | 0.055 | boundary | boundary|NMD3; | 0.0 |
| 2 | fp_D | Q96D46 | 75 | SRELLALCLKKIKAPLSK | CHHHHHHHHHHHCCCCCC | c4-4 | multi | 0.062 | 0.033 | 0.025 | boundary | MID|NMD3; | 0.0 |
| 3 | fp_D | Q96D46 | 79 | LALCLKKIKAPLSKVR | HHHHHHHHCCCCCCCC | c1aR-5 | multi | 0.069 | 0.033 | 0.032 | boundary | MID|NMD3; | 0.0 |
| 4 | fp_D | Q96D46 | 82 | CLKKIKAPLSKVRLVD | HHHHHCCCCCCCCEEE | c1a-5 | multi-selected | 0.084 | 0.032 | 0.048 | boundary | MID|NMD3; | 0.0 |
| 5 | fp_D | Q96D46 | 84 | KKIKAPLSKVRLVD | HHHCCCCCCCCEEE | c2-AT-4 | multi | 0.09 | 0.034 | 0.05 | boundary | MID|NMD3; | 0.0 |
| 6 | fp_beta_O | Q96D46 | 117 | QKEVMNGAILQQVFVVD | EEEEECCEEEEEEEEEE | c1c-4 | multi-selected | 0.026 | 0.022 | 0.041 | ORD | MID|NMD3; | 0.75 |
| 7 | fp_beta_O | Q96D46 | 120 | VMNGAILQQVFVVD | EECCEEEEEEEEEE | c2-AT-5 | multi | 0.028 | 0.022 | 0.027 | ORD | MID|NMD3; | 0.857 |
| 8 | fp_beta_O | Q96D46 | 128 | QVFVVDYVVQSQMCGDC | EEEEEEEEEEEECCCCC | c1cR-4 | uniq | 0.012 | 0.027 | 0.031 | ORD | MID|NMD3; | 1.0 |
| 9 | fp_O | Q96D46 | 164 | LHKKTFYYLEQLILKY | CHHHHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.048 | 0.046 | 0.033 | ORD | MID|NMD3; | 0.0 |
| 10 | fp_O | Q96D46 | 165 | HKKTFYYLEQLILKY | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.047 | 0.042 | 0.031 | ORD | MID|NMD3; | 0.0 |
| 11 | fp_beta_O | Q96D46 | 184 | NTLRIKEIHDGLDFYY | CCCCEEEECCEEEEEE | c1aR-4 | multi-selected | 0.013 | 0.034 | 0.114 | ORD | MID|NMD3; | 0.714 |
| 12 | fp_beta_O | Q96D46 | 184 | NTLRIKEIHDGLDFYY | CCCCEEEECCEEEEEE | c1d-4 | multi | 0.013 | 0.034 | 0.114 | ORD | MID|NMD3; | 0.714 |
| 13 | fp_O | Q96D46 | 261 | LAQSLGNMNQICVCI | HHHHHCCCCCEEEEE | c1b-4 | multi-selected | 0.024 | 0.031 | 0.049 | ORD | boundary|NMD3; | 0.143 |
| 14 | fp_beta_O | Q96D46 | 264 | SLGNMNQICVCIRVTS | HHCCCCCEEEEEEECC | c1d-5 | multi | 0.019 | 0.025 | 0.04 | ORD | boundary|NMD3; | 0.571 |
| 15 | fp_beta_O | Q96D46 | 269 | NQICVCIRVTSAIHLID | CCEEEEEEECCEEEEEC | c1c-4 | multi-selected | 0.012 | 0.013 | 0.047 | ORD | boundary|NMD3; | 0.75 |
| 16 | fp_beta_O | Q96D46 | 271 | ICVCIRVTSAIHLID | EEEEEEECCEEEEEC | c1b-AT-4 | multi | 0.011 | 0.01 | 0.046 | ORD | boundary|NMD3; | 0.714 |
| 17 | fp_O | Q96D46 | 280 | AIHLIDPNTLQVAD | EEEEECCCCCCEEE | c3-AT | uniq | 0.017 | 0.035 | 0.123 | ORD | boundary|NMD3; | 0.286 |
| 18 | fp_beta_D | Q96D46 | 315 | EFIVMECSIVQDIKRAA | EEEEEEEEEEECCCCCC | c1cR-4 | uniq | 0.096 | 0.142 | 0.12 | boundary | 0.875 | |
| 19 | cand_D | Q96D46 | 476 | PRISLAEMLEDLHISQ .*..*...*..*.*.. | CCCCHHHHHHCCCCCC | c1aR-4 | multi-selected | 0.703 | 0.43 | 0.578 | DISO | 0.0 | |
| 20 | cand_D | Q96D46 | 476 | PRISLAEMLEDLHISQ .*..*...*..*.*.. | CCCCHHHHHHCCCCCC | c1a-4 | multi-selected | 0.703 | 0.43 | 0.578 | DISO | 0.0 | |
| 21 | cand_D | Q96D46 | 476 | PRISLAEMLEDLHISQ .*..*...*..*.*.. | CCCCHHHHHHCCCCCC | c1d-4 | multi | 0.703 | 0.43 | 0.578 | DISO | 0.0 | |
| 22 | cand_D | Q96D46 | 477 | RISLAEMLEDLHISQ *..*...*..*.*.. | CCCHHHHHHCCCCCC | c1b-AT-5 | multi | 0.702 | 0.431 | 0.573 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment