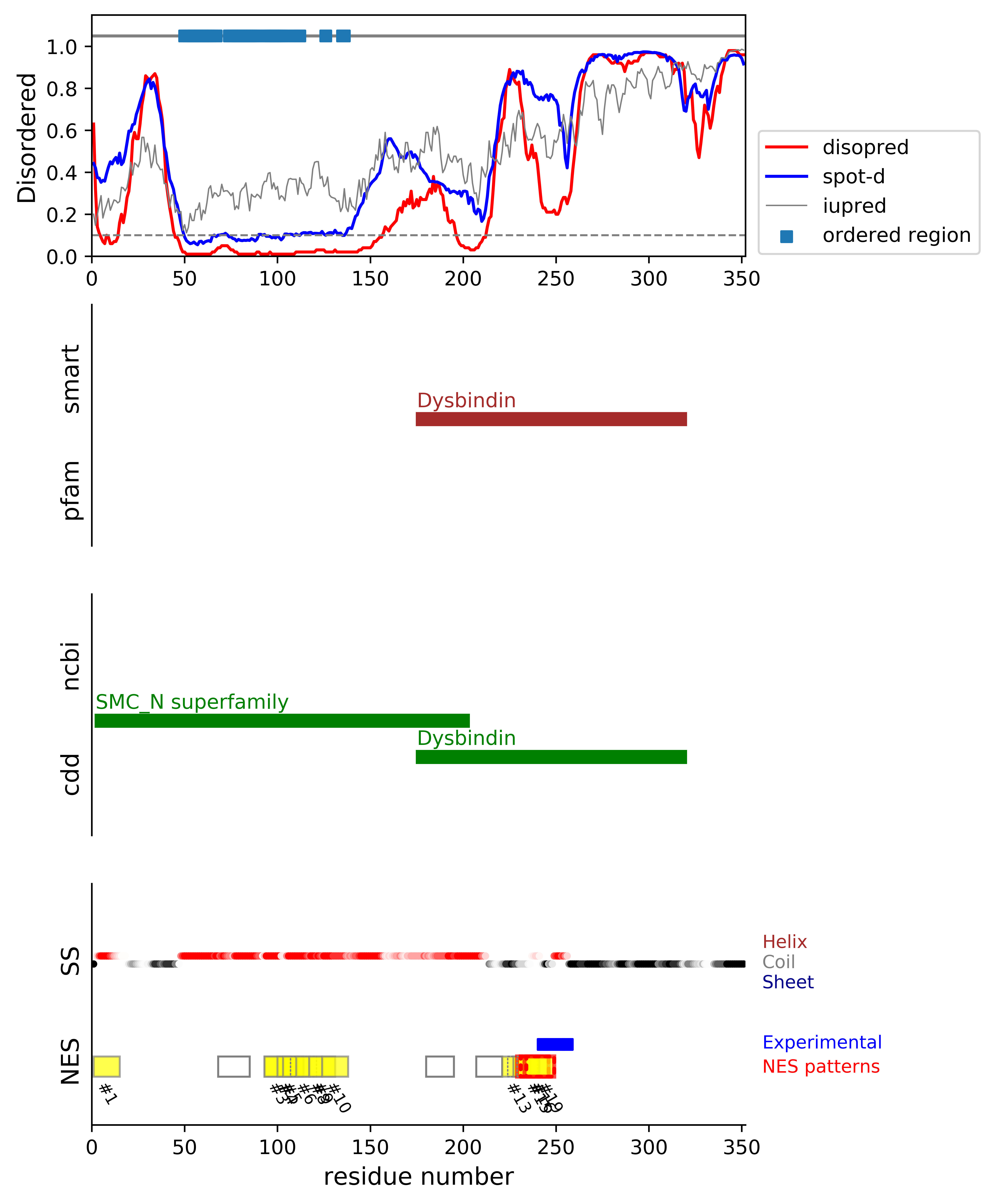

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q96EV8 | 1 | MLETLRERLLSVQQ | CHHHHHHHHHHHHH | c3-4 | uniq | 0.14 | 0.412 | 0.232 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 2 | fp_D | Q96EV8 | 68 | DCASAGELVDSEVVMLS | HHHHHHHHHHHHHHHHH | c1c-AT-4 | uniq | 0.028 | 0.088 | 0.295 | boundary | MID|SMC_N superfamily; | 0.0 |
| 3 | fp_D | Q96EV8 | 93 | SLVELQEQLQQLPALIAD | HHHHHHHHHHHHHHHHHH | c4-5 | multi-selected | 0.011 | 0.096 | 0.339 | boundary | MID|SMC_N superfamily; | 0.0 |
| 4 | fp_D | Q96EV8 | 93 | SLVELQEQLQQLPA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.011 | 0.094 | 0.331 | boundary | MID|SMC_N superfamily; | 0.0 |
| 5 | fp_D | Q96EV8 | 100 | QLQQLPALIADLES | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.013 | 0.099 | 0.33 | boundary | MID|SMC_N superfamily; | 0.0 |
| 6 | fp_D | Q96EV8 | 103 | QLPALIADLESMTA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.015 | 0.104 | 0.334 | boundary | MID|SMC_N superfamily; | 0.0 |
| 7 | fp_D | Q96EV8 | 107 | LIADLESMTANLTH | HHHHHHHHHHHHHH | c3-AT | multi | 0.018 | 0.107 | 0.353 | boundary | MID|SMC_N superfamily; | 0.0 |
| 8 | fp_D | Q96EV8 | 110 | DLESMTANLTHLEA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.022 | 0.108 | 0.365 | boundary | MID|SMC_N superfamily; | 0.0 |
| 9 | fp_D | Q96EV8 | 117 | NLTHLEASFEEVEN | HHHHHHHHHHHHHH | c3-4 | uniq | 0.024 | 0.11 | 0.362 | boundary | MID|SMC_N superfamily; | 0.0 |
| 10 | fp_D | Q96EV8 | 124 | SFEEVENNLLHLED | HHHHHHHHHHHHHH | c3-4 | uniq | 0.022 | 0.109 | 0.292 | DISO | MID|SMC_N superfamily; | 0.0 |
| 11 | fp_D | Q96EV8 | 180 | KVLEMEHTQQMKLKE | HHHHHHHHHHHHHHH | c1b-AT-5 | uniq | 0.273 | 0.344 | 0.515 | DISO | boundary|SMC_N superfamily; boundary|Dysbindin; | 0.0 |
| 12 | fp_D | Q96EV8 | 207 | MEQYLSTGYLQIAE | HHHHHHCCCCCCCC | c2-AT-4 | uniq | 0.236 | 0.369 | 0.412 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 13 | fp_D | Q96EV8 | 221 | RREPIGSMSSMEVNV | CCCCCCCCCCCCCCC | c1b-4 | multi-selected | 0.733 | 0.84 | 0.587 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 14 | fp_D | Q96EV8 | 224 | PIGSMSSMEVNVDMLE | CCCCCCCCCCCCCHHH | c1d-5 | multi | 0.678 | 0.838 | 0.593 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 15 | fp_D | Q96EV8 | 227 | SMSSMEVNVDMLEQ | CCCCCCCCCCHHHH | c3-4 | multi-selected | 0.621 | 0.833 | 0.611 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 16 | cand_D | Q96EV8 | 229 | SSMEVNVDMLEQMDLMD * | CCCCCCCCHHHHHCCCC | c1c-4 | multi-selected | 0.488 | 0.806 | 0.598 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 17 | fp_D | Q96EV8 | 229 | SSMEVNVDMLEQMDLMD * | CCCCCCCCHHHHHCCCC | c1cR-5 | multi | 0.488 | 0.806 | 0.598 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 18 | cand_D | Q96EV8 | 231 | MEVNVDMLEQMDLMD * | CCCCCCHHHHHCCCC | c1b-4 | multi | 0.443 | 0.797 | 0.587 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 19 | cand_D | Q96EV8 | 233 | VNVDMLEQMDLMDISD * * | CCCCHHHHHCCCCCCC | c1a-4 | multi-selected | 0.366 | 0.78 | 0.555 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
| 20 | cand_D | Q96EV8 | 234 | NVDMLEQMDLMDISD * * | CCCHHHHHCCCCCCC | c1b-5 | multi | 0.351 | 0.777 | 0.551 | DISO | boundary|SMC_N superfamily; MID|Dysbindin; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment