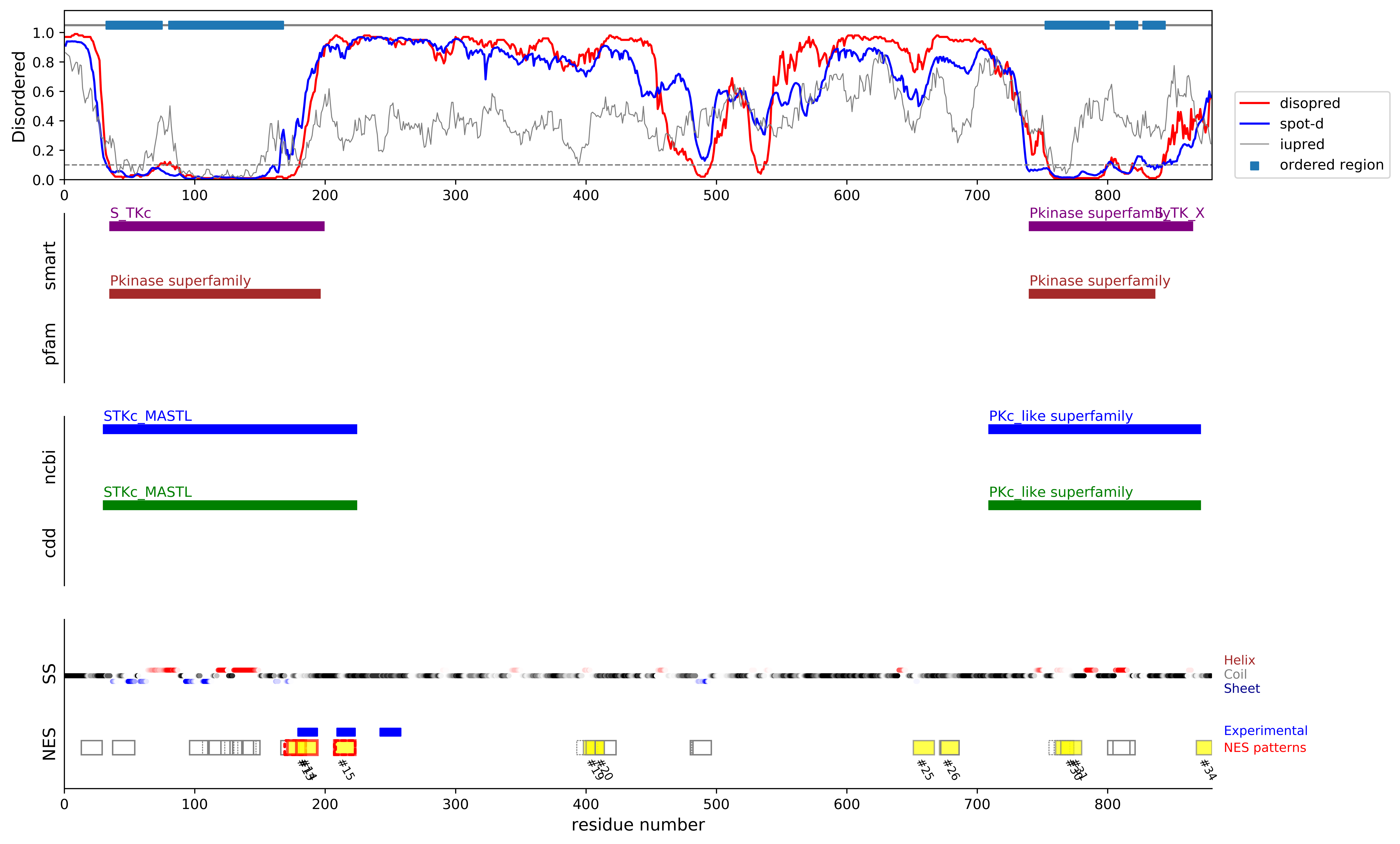

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q96GX5 | 13 | GGAATEEGVNRIAVPK | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.911 | 0.716 | 0.523 | boundary | boundary|STKc_MASTL; | 0.0 |
| 2 | fp_D | Q96GX5 | 37 | IVKPISRGAFGKVYLGQ | EEEECCCCCCCEEEEEE | c1c-AT-5 | uniq | 0.023 | 0.039 | 0.117 | boundary | boundary|STKc_MASTL; | 0.125 |
| 3 | fp_beta_D | Q96GX5 | 96 | LYYSLQSANNVYLVM | EEEEEECCCEEEEEE | c1b-AT-4 | uniq | 0.015 | 0.01 | 0.035 | __ | MID|STKc_MASTL; | 0.571 |
| 4 | fp_beta_O | Q96GX5 | 106 | VYLVMEYLIGGDVKSLL | EEEEEEEECCCCHHHHH | c1cR-5 | multi | 0.01 | 0.012 | 0.032 | ORD | MID|STKc_MASTL; | 0.5 |
| 5 | fp_O | Q96GX5 | 110 | MEYLIGGDVKSLLHIYG | EEEECCCCHHHHHHHHC | c1c-4 | multi-selected | 0.01 | 0.014 | 0.033 | ORD | MID|STKc_MASTL; | 0.0 |
| 6 | fp_O | Q96GX5 | 120 | SLLHIYGYFDEEMAVKY | HHHHHHCCCCHHHHHHH | c1c-5 | uniq | 0.01 | 0.014 | 0.029 | ORD | MID|STKc_MASTL; | 0.0 |
| 7 | fp_O | Q96GX5 | 129 | DEEMAVKYISEVALAL | CHHHHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.01 | 0.01 | 0.031 | ORD | MID|STKc_MASTL; | 0.0 |
| 8 | fp_O | Q96GX5 | 130 | EEMAVKYISEVALAL | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.01 | 0.009 | 0.031 | ORD | MID|STKc_MASTL; | 0.0 |
| 9 | fp_O | Q96GX5 | 133 | AVKYISEVALALDY | HHHHHHHHHHHHHH | c3-AT | multi | 0.01 | 0.008 | 0.036 | ORD | MID|STKc_MASTL; | 0.0 |
| 10 | fp_O | Q96GX5 | 136 | YISEVALALDYLHR | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.009 | 0.049 | ORD | MID|STKc_MASTL; | 0.0 |
| 11 | fp_beta_D | Q96GX5 | 166 | NEGHIKLTDFGLSK | CCCCEEEEECCCCC | c2-4 | multi-selected | 0.028 | 0.253 | 0.194 | boundary | MID|STKc_MASTL; | 0.714 |
| 12 | cand_D | Q96GX5 | 169 | HIKLTDFGLSKVTLNR * | CEEEEECCCCCCCCCC | c1a-AT-5 | multi | 0.079 | 0.293 | 0.165 | boundary | MID|STKc_MASTL; | 0.286 |
| 13 | cand_D | Q96GX5 | 171 | KLTDFGLSKVTLNR * | EEEECCCCCCCCCC | c2-5 | multi-selected | 0.089 | 0.303 | 0.158 | boundary | MID|STKc_MASTL; | 0.143 |
| 14 | cand_D | Q96GX5 | 178 | SKVTLNRDINMMDILT * * * * | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.351 | 0.567 | 0.224 | boundary | MID|STKc_MASTL; | 0.0 |
| 15 | cand_D | Q96GX5 | 207 | TPGQVLSLISSLGFNT * * * * | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.942 | 0.889 | 0.29 | DISO | boundary|STKc_MASTL; | 0.0 |
| 16 | cand_D | Q96GX5 | 207 | TPGQVLSLISSLGFNT * * * * | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.942 | 0.889 | 0.29 | DISO | boundary|STKc_MASTL; | 0.0 |
| 17 | cand_D | Q96GX5 | 208 | PGQVLSLISSLGFNT * * * * | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.94 | 0.889 | 0.283 | DISO | boundary|STKc_MASTL; | 0.0 |
| 18 | fp_D | Q96GX5 | 393 | FSGEVSWEAVELDVNN | CCCCCHHHHHHCCCCC | c1a-AT-4 | multi | 0.841 | 0.75 | 0.282 | DISO | 0.0 | |
| 19 | fp_D | Q96GX5 | 398 | SWEAVELDVNNINMDT | HHHHHHCCCCCCCCCC | c1a-4 | multi-selected | 0.863 | 0.791 | 0.373 | DISO | 0.0 | |
| 20 | fp_D | Q96GX5 | 400 | EAVELDVNNINMDT | HHHHCCCCCCCCCC | c2-4 | multi-selected | 0.869 | 0.8 | 0.395 | DISO | 0.0 | |
| 21 | fp_D | Q96GX5 | 407 | NNINMDTDTSQLGFHQ | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.929 | 0.873 | 0.468 | DISO | 0.0 | |
| 22 | fp_beta_D | Q96GX5 | 480 | QNTGLTVEVQDLKLSV | CCCCCEEEEEEEEEEC | c1a-4 | multi-selected | 0.058 | 0.25 | 0.403 | DISO | 0.857 | |
| 23 | fp_beta_D | Q96GX5 | 481 | NTGLTVEVQDLKLSV | CCCCEEEEEEEEEEC | c1b-AT-4 | multi | 0.053 | 0.229 | 0.398 | DISO | 1.0 | |
| 24 | fp_beta_D | Q96GX5 | 482 | TGLTVEVQDLKLSV | CCCEEEEEEEEEEC | c2-4 | multi-selected | 0.047 | 0.211 | 0.398 | DISO | 1.0 | |
| 25 | fp_D | Q96GX5 | 651 | NAVAFRSFNSHINASN | CCEECCCCCCCCCCCC | c1aR-4 | uniq | 0.822 | 0.618 | 0.501 | DISO | 0.0 | |
| 26 | fp_D | Q96GX5 | 671 | SRMNMTSLDAMDISC | CCCCCCCCCCCCCCC | c1b-4 | multi-selected | 0.959 | 0.805 | 0.487 | DISO | 0.0 | |
| 27 | fp_D | Q96GX5 | 672 | RMNMTSLDAMDISC | CCCCCCCCCCCCCC | c2-AT-5 | multi-selected | 0.958 | 0.803 | 0.472 | DISO | 0.0 | |
| 28 | fp_D | Q96GX5 | 755 | HGPAVDWWALGVCLFE | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi | 0.018 | 0.026 | 0.098 | boundary | MID|PKc_like superfamily; | 0.0 |
| 29 | fp_D | Q96GX5 | 759 | VDWWALGVCLFEFLT | CCHHHHHHHHHHHCC | c1b-AT-4 | multi | 0.01 | 0.017 | 0.087 | boundary | MID|PKc_like superfamily; | 0.0 |
| 30 | fp_D | Q96GX5 | 760 | DWWALGVCLFEFLT | CHHHHHHHHHHHCC | c2-4 | multi-selected | 0.01 | 0.016 | 0.089 | boundary | MID|PKc_like superfamily; | 0.0 |
| 31 | fp_D | Q96GX5 | 764 | LGVCLFEFLTGIPPFN | HHHHHHHHCCCCCCCC | c1aR-5 | multi-selected | 0.01 | 0.018 | 0.179 | boundary | MID|PKc_like superfamily; | 0.0 |
| 32 | fp_D | Q96GX5 | 800 | GEEKLSDNAQSAVEILL | CCCCCCHHHHHHHHHHC | c1c-AT-4 | multi-selected | 0.086 | 0.075 | 0.469 | boundary | MID|PKc_like superfamily; | 0.0 |
| 33 | fp_D | Q96GX5 | 804 | LSDNAQSAVEILLTIDD | CCHHHHHHHHHHCCCCC | c1c-AT-4 | multi-selected | 0.077 | 0.064 | 0.41 | boundary | MID|PKc_like superfamily; | 0.0 |
| 34 | fp_D | Q96GX5 | 868 | TAQHLTVSGFSLXX | CCCCCCCCCCCC | c2-4-Ct | uniq | 0.425 | 0.458 | 0.325 | DISO | boundary|PKc_like superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment