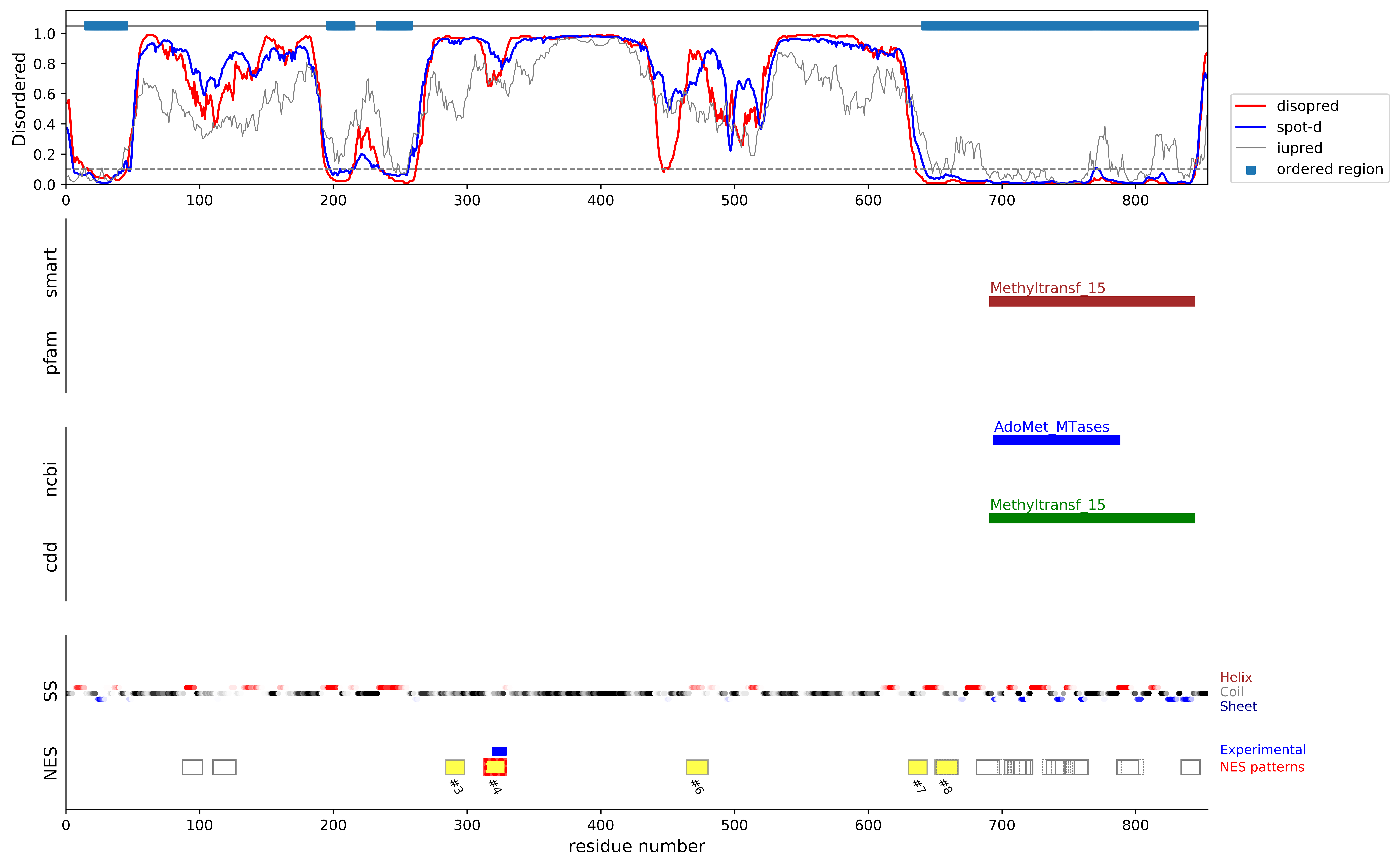

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q96RS0 | 87 | LDSEAELMRSMGLPL | CCHHHHHHHHCCCCC | c1b-AT-4 | uniq | 0.645 | 0.796 | 0.43 | DISO | 0.0 | |
| 2 | fp_D | Q96RS0 | 110 | KDFEVSMNTRNKVKIKK | CCCEECCCCCCCCHHHH | c1c-AT-4 | uniq | 0.563 | 0.766 | 0.413 | DISO | 0.125 | |
| 3 | fp_D | Q96RS0 | 284 | DEKCMKVDLVSFPS | CCCCCCCCCCCCCC | c2-4 | uniq | 0.973 | 0.947 | 0.494 | DISO | 0.0 | |
| 4 | cand_D | Q96RS0 | 313 | DHSEILDGISNIKLNS * * * | CCCCCCCCCCCCCCCH | c1a-4 | multi-selected | 0.734 | 0.862 | 0.696 | DISO | 0.0 | |
| 5 | cand_D | Q96RS0 | 314 | HSEILDGISNIKLNS * * * | CCCCCCCCCCCCCCH | c1b-4 | multi | 0.726 | 0.856 | 0.688 | DISO | 0.0 | |
| 6 | fp_D | Q96RS0 | 464 | SHRQVRYLEKNVKLKS | CCCHHHHHHHHHHCCC | c1d-4 | uniq | 0.819 | 0.72 | 0.491 | DISO | 0.0 | |
| 7 | fp_D | Q96RS0 | 630 | KVNGLPPEIAAVPE | CCCCCCCCCCCCHH | c3-4 | uniq | 0.143 | 0.306 | 0.437 | boundary | 0.0 | |
| 8 | fp_D | Q96RS0 | 650 | QRYRLFSRFDDGIKLDR | HHHHHHCCCCCCCCCCC | c1c-4 | multi-selected | 0.017 | 0.05 | 0.135 | boundary | 0.0 | |
| 9 | fp_D | Q96RS0 | 651 | RYRLFSRFDDGIKLDR | HHHHHCCCCCCCCCCC | c1d-4 | multi | 0.018 | 0.051 | 0.135 | boundary | 0.0 | |
| 10 | fp_O | Q96RS0 | 681 | IAGRVSQSFKCDVVVDA | HHHHHHCCCCCCEEEEE | c1c-4 | uniq | 0.013 | 0.018 | 0.121 | ORD | boundary|Methyltransf_15; | 0.0 |
| 11 | fp_O | Q96RS0 | 697 | AFCGVGGNTIQFALTG | ECCCCCCCHHHHHHCC | c1a-AT-5 | multi | 0.01 | 0.011 | 0.055 | ORD | boundary|Methyltransf_15; | 0.0 |
| 12 | fp_O | Q96RS0 | 702 | GGNTIQFALTGMRVIA | CCCHHHHHHCCCEEEE | c1a-4 | multi-selected | 0.01 | 0.009 | 0.054 | ORD | boundary|Methyltransf_15; | 0.0 |
| 13 | fp_O | Q96RS0 | 702 | GGNTIQFALTGMRVIA | CCCHHHHHHCCCEEEE | c1d-AT-4 | multi | 0.01 | 0.009 | 0.054 | ORD | boundary|Methyltransf_15; | 0.0 |
| 14 | fp_O | Q96RS0 | 704 | NTIQFALTGMRVIA | CHHHHHHCCCEEEE | c2-4 | multi-selected | 0.01 | 0.008 | 0.055 | ORD | boundary|Methyltransf_15; | 0.0 |
| 15 | fp_O | Q96RS0 | 705 | TIQFALTGMRVIAIDI | HHHHHHCCCEEEEEEC | c1a-AT-5 | multi | 0.01 | 0.007 | 0.05 | ORD | boundary|Methyltransf_15; | 0.286 |
| 16 | fp_O | Q96RS0 | 706 | IQFALTGMRVIAIDI | HHHHHCCCEEEEEEC | c1b-4 | multi | 0.01 | 0.007 | 0.049 | ORD | boundary|Methyltransf_15; | 0.429 |
| 17 | fp_beta_O | Q96RS0 | 707 | QFALTGMRVIAIDIDP | HHHHCCCEEEEEECCH | c1a-AT-5 | multi | 0.01 | 0.007 | 0.052 | ORD | boundary|Methyltransf_15; | 0.571 |
| 18 | fp_O | Q96RS0 | 707 | QFALTGMRVIAIDI | HHHHCCCEEEEEEC | c2-AT-5 | multi | 0.01 | 0.007 | 0.048 | ORD | boundary|Methyltransf_15; | 0.429 |
| 19 | fp_beta_O | Q96RS0 | 709 | ALTGMRVIAIDIDP | HHCCCEEEEEECCH | c2-5 | multi-selected | 0.01 | 0.007 | 0.055 | ORD | boundary|Methyltransf_15; | 0.714 |
| 20 | fp_beta_O | Q96RS0 | 709 | ALTGMRVIAIDIDP | HHCCCEEEEEECCH | c3-AT | multi | 0.01 | 0.007 | 0.055 | ORD | boundary|Methyltransf_15; | 0.714 |
| 21 | fp_O | Q96RS0 | 730 | NNAEVYGIADKIEFIC | HHHHHHCCCCCEEEEE | c1a-AT-4 | multi | 0.01 | 0.013 | 0.036 | ORD | MID|Methyltransf_15; | 0.0 |
| 22 | fp_O | Q96RS0 | 730 | NNAEVYGIADKIEFIC | HHHHHHCCCCCEEEEE | c1d-4 | multi | 0.01 | 0.013 | 0.036 | ORD | MID|Methyltransf_15; | 0.0 |
| 23 | fp_O | Q96RS0 | 733 | EVYGIADKIEFICG | HHHCCCCCEEEEEC | c3-4 | multi-selected | 0.01 | 0.013 | 0.022 | ORD | MID|Methyltransf_15; | 0.286 |
| 24 | fp_beta_O | Q96RS0 | 737 | IADKIEFICGDFLLLA | CCCCEEEEECHHHHHH | c1d-4 | multi | 0.01 | 0.011 | 0.014 | ORD | MID|Methyltransf_15; | 0.714 |
| 25 | fp_O | Q96RS0 | 740 | KIEFICGDFLLLAS | CEEEEECHHHHHHH | c3-4 | multi-selected | 0.01 | 0.01 | 0.008 | ORD | MID|Methyltransf_15; | 0.429 |
| 26 | fp_O | Q96RS0 | 747 | DFLLLASFLKADVVFLS | HHHHHHHHCCCCEEEEC | c1c-5 | multi-selected | 0.01 | 0.013 | 0.019 | ORD | MID|Methyltransf_15; | 0.0 |
| 27 | fp_O | Q96RS0 | 748 | FLLLASFLKADVVFLS | HHHHHHHCCCCEEEEC | c1d-AT-5 | multi | 0.01 | 0.014 | 0.019 | ORD | MID|Methyltransf_15; | 0.0 |
| 28 | fp_O | Q96RS0 | 750 | LLASFLKADVVFLSP | HHHHHCCCCEEEECC | c1b-AT-5 | multi | 0.01 | 0.014 | 0.023 | ORD | MID|Methyltransf_15; | 0.286 |
| 29 | fp_O | Q96RS0 | 751 | LASFLKADVVFLSP | HHHHCCCCEEEECC | c2-AT-4 | multi | 0.01 | 0.015 | 0.025 | ORD | MID|Methyltransf_15; | 0.286 |
| 30 | fp_O | Q96RS0 | 786 | DGFEIFRLSKKITNNI | CHHHHHHHHHHHCCCE | c1aR-4 | multi-selected | 0.01 | 0.011 | 0.065 | ORD | MID|Methyltransf_15; | 0.0 |
| 31 | fp_O | Q96RS0 | 789 | EIFRLSKKITNNIVYFL | HHHHHHHHHCCCEEEEC | c1cR-5 | multi | 0.01 | 0.009 | 0.063 | ORD | MID|Methyltransf_15; | 0.0 |
| 32 | fp_beta_D | Q96RS0 | 834 | KLKTITAYFGDLIR | EEEEEEEEECCCCC | c3-4 | uniq | 0.054 | 0.06 | 0.104 | boundary | boundary|Methyltransf_15; | 0.857 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment