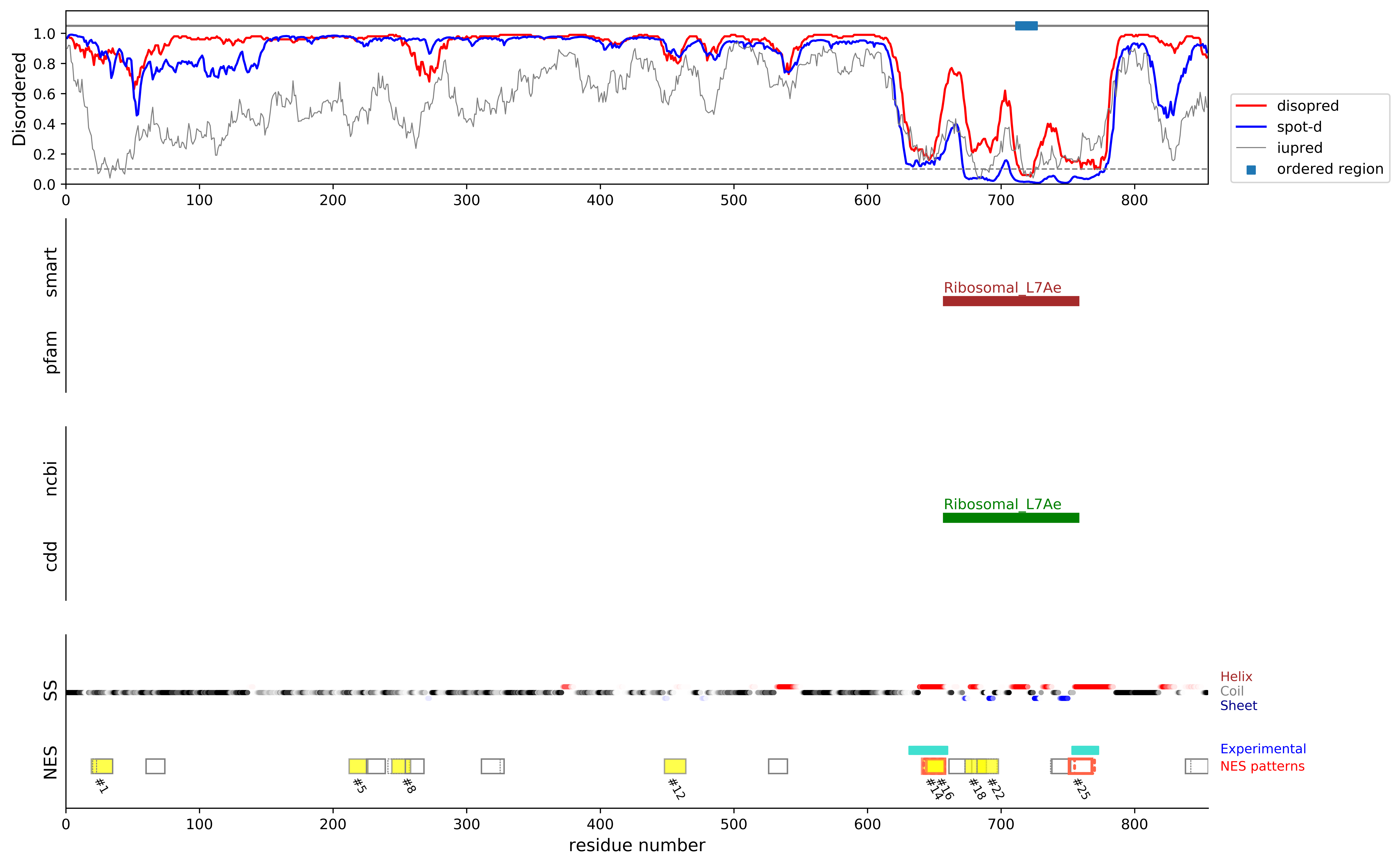

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q96T21 | 19 | DVKPFVPRFAGLNVAW | CCCCCCCCCCCCCCCC | c1a-5 | multi-selected | 0.847 | 0.875 | 0.139 | DISO | 0.0 | |
| 2 | fp_D | Q96T21 | 20 | VKPFVPRFAGLNVAW | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.845 | 0.87 | 0.127 | DISO | 0.0 | |
| 3 | fp_D | Q96T21 | 23 | FVPRFAGLNVAW | CCCCCCCCCCCC | c2-rev | multi | 0.848 | 0.853 | 0.107 | DISO | 0.0 | |
| 4 | fp_D | Q96T21 | 60 | TEQKIYTEDMAFGA | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.884 | 0.763 | 0.369 | DISO | 0.0 | |
| 5 | fp_D | Q96T21 | 212 | SKPEFEFTTLDFPE | CCCCCCCCCCCCCC | c2-4 | uniq | 0.968 | 0.95 | 0.438 | DISO | 0.0 | |
| 6 | fp_D | Q96T21 | 225 | ELQGAENNMSEIQK | CCCCCCCCCCCCCC | c3-AT | uniq | 0.979 | 0.953 | 0.593 | DISO | 0.0 | |
| 7 | fp_D | Q96T21 | 241 | KWGPVHSVSTDISLLR | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.966 | 0.962 | 0.443 | DISO | 0.0 | |
| 8 | fp_D | Q96T21 | 244 | PVHSVSTDISLLRE | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.958 | 0.96 | 0.406 | DISO | 0.0 | |
| 9 | fp_D | Q96T21 | 254 | LLREVVKPAAVLSK | CCCCCCCCCCCCCC | c3-AT | uniq | 0.846 | 0.959 | 0.355 | DISO | 0.0 | |
| 10 | fp_D | Q96T21 | 311 | ELSAAPKNVTSMINLKT | CCCCCCCCCCCCCCCCC | c1c-AT-5 | multi-selected | 0.982 | 0.964 | 0.529 | DISO | 0.0 | |
| 11 | fp_D | Q96T21 | 311 | ELSAAPKNVTSMIN | CCCCCCCCCCCCCC | c3-AT | multi | 0.98 | 0.966 | 0.515 | DISO | 0.0 | |
| 12 | fp_D | Q96T21 | 448 | VQLDLGGMLTALEKKQ | EEECCCCCHHHHHHHH | c1aR-4 | uniq | 0.848 | 0.879 | 0.656 | DISO | 0.0 | |
| 13 | fp_D | Q96T21 | 526 | AKKPTSLKKIILKE | CCCCCHHHHHHHHH | c2-AT-4 | uniq | 0.871 | 0.856 | 0.67 | DISO | 0.0 | |
| 14 | cand_D | Q96T21 | 641 | VDACVTDLLKELVRFQD +++++++++++++++++ | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.279 | 0.158 | 0.217 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 15 | cand_D | Q96T21 | 642 | DACVTDLLKELVRFQD ++++++++++++++++ | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.282 | 0.159 | 0.217 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 16 | cand_D | Q96T21 | 644 | CVTDLLKELVRFQD ++++++++++++++ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.296 | 0.163 | 0.224 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 17 | fp_D | Q96T21 | 661 | QKDPVKAKTKRRLVLGL | CCCHHHHCCCCEEEEHH | c1c-AT-4 | uniq | 0.621 | 0.23 | 0.325 | DISO | boundary|Ribosomal_L7Ae; | 0.125 |
| 18 | fp_D | Q96T21 | 673 | LVLGLREVLKHLKLKK | EEEHHHHHHHHHHCCC | c1a-5 | multi-selected | 0.297 | 0.04 | 0.135 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 19 | fp_D | Q96T21 | 673 | LVLGLREVLKHLKLKK | EEEHHHHHHHHHHCCC | c1aR-4 | multi | 0.297 | 0.04 | 0.135 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 20 | fp_D | Q96T21 | 673 | LVLGLREVLKHLKLKK | EEEHHHHHHHHHHCCC | c1d-5 | multi | 0.297 | 0.04 | 0.135 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 21 | fp_D | Q96T21 | 682 | KHLKLKKLKCVIISP | HHHHCCCCEEEEECC | c1b-4 | multi | 0.27 | 0.033 | 0.098 | DISO | MID|Ribosomal_L7Ae; | 0.429 |
| 22 | fp_D | Q96T21 | 682 | KHLKLKKLKCVIISPN | HHHHCCCCEEEEECCC | c1aR-4 | multi-selected | 0.271 | 0.034 | 0.104 | DISO | MID|Ribosomal_L7Ae; | 0.429 |
| 23 | fp_beta_D | Q96T21 | 737 | SLNKAVPVSVVGIFS | HHCCCCCEEEEEEEC | c1b-AT-5 | multi | 0.259 | 0.025 | 0.169 | boundary | boundary|Ribosomal_L7Ae; | 0.571 |
| 24 | fp_beta_D | Q96T21 | 738 | LNKAVPVSVVGIFS | HCCCCCEEEEEEEC | c2-4 | multi-selected | 0.249 | 0.023 | 0.17 | boundary | boundary|Ribosomal_L7Ae; | 0.571 |
| 25 | cand_D | Q96T21 | 751 | SYDGAQDQFHKMVELTV ++++++++++++ | CCCCHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.145 | 0.038 | 0.222 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 26 | cand_D | Q96T21 | 755 | AQDQFHKMVELTVAA ++++++++++++++ | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.145 | 0.041 | 0.238 | DISO | boundary|Ribosomal_L7Ae; | 0.0 |
| 27 | fp_D | Q96T21 | 838 | LEESLEASTSQMMNLNL | HHHHHHHHHHHHHCCCC | c1c-AT-4 | multi-selected | 0.941 | 0.879 | 0.509 | DISO | 0.0 | |
| 28 | fp_D | Q96T21 | 842 | LEASTSQMMNLNLXX | HHHHHHHHHCCCC | c1b-AT-4-Ct | multi | 0.929 | 0.904 | 0.524 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment