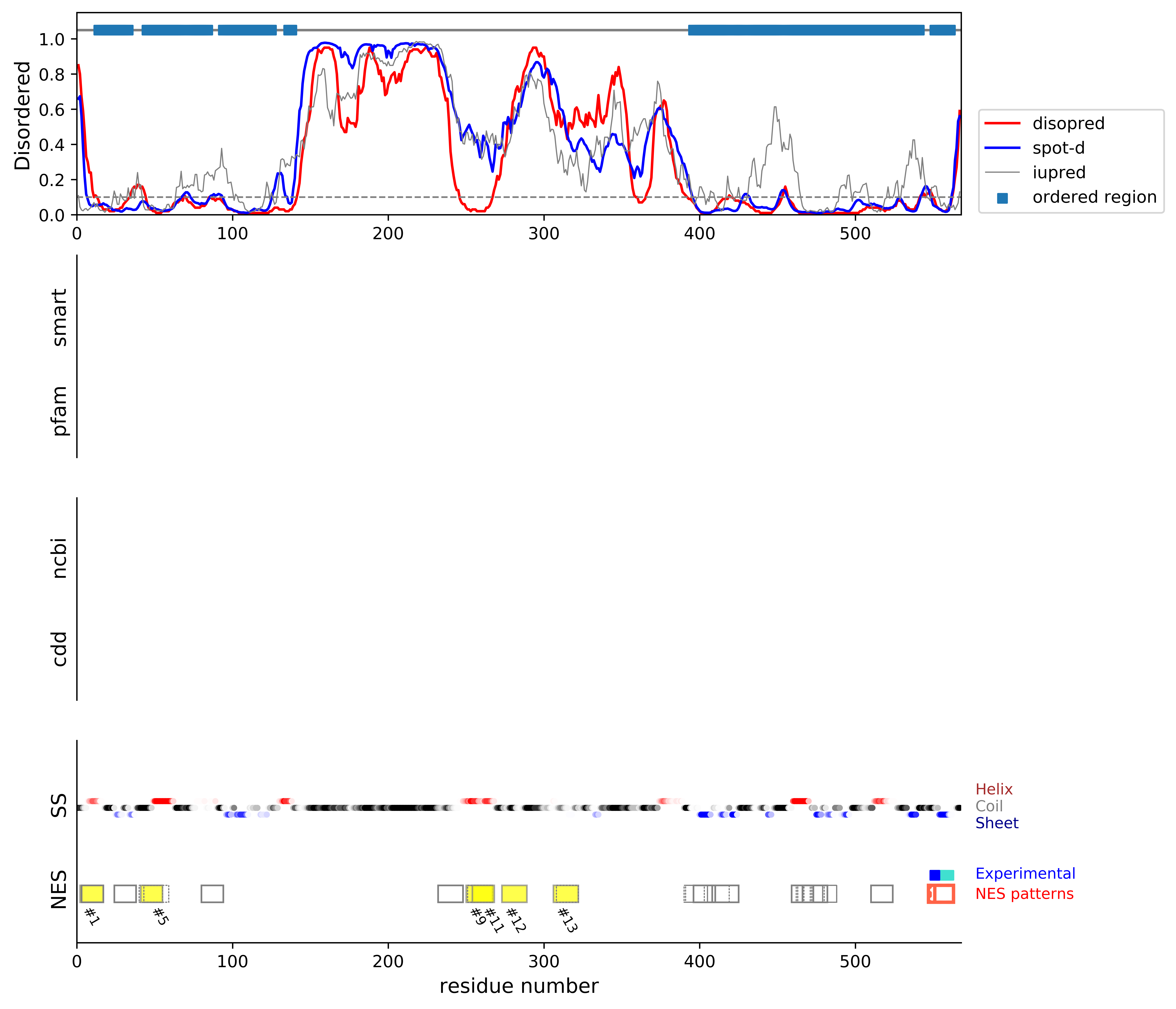

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q99AM3 | 2 | AGRRLTWISEFIVGA | CCCCCHHHHHHHHHH | c1b-4 | multi-selected | 0.285 | 0.169 | 0.038 | boundary | 0.0 | |
| 2 | fp_D | Q99AM3 | 3 | GRRLTWISEFIVGA | CCCCHHHHHHHHHH | c2-AT-4 | multi-selected | 0.248 | 0.133 | 0.038 | boundary | 0.0 | |
| 3 | fp_D | Q99AM3 | 24 | LVKWLDRSTGTFLA | CEEEEECCCCEEEE | c3-AT | uniq | 0.074 | 0.027 | 0.101 | boundary | 0.429 | |
| 4 | fp_D | Q99AM3 | 40 | ARNDVIPLDSLQFFI | CCCCCCCCCHHHHHH | c1b-4 | multi | 0.065 | 0.048 | 0.055 | boundary | 0.0 | |
| 5 | fp_D | Q99AM3 | 41 | RNDVIPLDSLQFFI | CCCCCCCCHHHHHH | c2-4 | multi-selected | 0.058 | 0.048 | 0.048 | boundary | 0.0 | |
| 6 | fp_D | Q99AM3 | 43 | DVIPLDSLQFFIDFKR | CCCCCCHHHHHHHHHH | c1d-5 | multi | 0.036 | 0.038 | 0.035 | boundary | 0.0 | |
| 7 | fp_D | Q99AM3 | 80 | KICTTSRRLRRLPG | HHHCCCHHHHCCCC | c3-AT | uniq | 0.079 | 0.095 | 0.232 | boundary | 0.0 | |
| 8 | fp_D | Q99AM3 | 232 | TTVHTDNVEDDLTLLD | CCCCCCCCCCCCCCHH | c1d-AT-4 | uniq | 0.449 | 0.703 | 0.74 | DISO | 0.0 | |
| 9 | fp_D | Q99AM3 | 250 | SACALMYHVGQEMDMLM | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.031 | 0.416 | 0.422 | DISO | 0.0 | |
| 10 | fp_D | Q99AM3 | 251 | ACALMYHVGQEMDMLM | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.031 | 0.413 | 0.419 | DISO | 0.0 | |
| 11 | fp_D | Q99AM3 | 254 | LMYHVGQEMDMLMR | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.036 | 0.382 | 0.417 | DISO | 0.0 | |
| 12 | fp_D | Q99AM3 | 273 | DLFDLLGIPEDVIATS | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.597 | 0.565 | 0.531 | DISO | 0.0 | |
| 13 | fp_D | Q99AM3 | 306 | AASAVGAGVEDVYLAG | HHCCCCCCCCEEECCC | c1a-4 | multi-selected | 0.537 | 0.534 | 0.326 | DISO | 0.143 | |
| 14 | fp_D | Q99AM3 | 308 | SAVGAGVEDVYLAG | CCCCCCCCEEECCC | c2-AT-4 | multi | 0.53 | 0.501 | 0.306 | DISO | 0.286 | |
| 15 | fp_D | Q99AM3 | 390 | ARPPVMITDRLGVEV | CCCCCCCCCCEEEEE | c1b-AT-4 | multi | 0.057 | 0.101 | 0.134 | boundary | 0.143 | |
| 16 | fp_D | Q99AM3 | 391 | RPPVMITDRLGVEV | CCCCCCCCCEEEEE | c2-AT-4 | multi | 0.052 | 0.088 | 0.129 | boundary | 0.143 | |
| 17 | fp_beta_D | Q99AM3 | 396 | ITDRLGVEVFYFGR | CCCCEEEEEEEECC | c2-4 | multi-selected | 0.024 | 0.027 | 0.107 | boundary | 0.857 | |
| 18 | fp_D | Q99AM3 | 403 | EVFYFGRPAMSLEVER | EEEEECCCCCEEEEEC | c1a-AT-5 | multi | 0.046 | 0.022 | 0.087 | boundary | 0.286 | |
| 19 | fp_beta_D | Q99AM3 | 408 | GRPAMSLEVERKVFILC | CCCCCEEEEECCEEEEE | c1c-4 | multi-selected | 0.073 | 0.026 | 0.093 | boundary | 0.625 | |
| 20 | fp_O | Q99AM3 | 459 | DVNLLAAVLRSFAS | CHHHHHHHHHHHCC | c3-4 | multi-selected | 0.022 | 0.021 | 0.136 | ORD | 0.0 | |
| 21 | fp_O | Q99AM3 | 462 | LLAAVLRSFASGLVIVS | HHHHHHHHHCCCCEEEE | c1c-5 | multi | 0.012 | 0.014 | 0.062 | ORD | 0.0 | |
| 22 | fp_O | Q99AM3 | 463 | LAAVLRSFASGLVIVSL | HHHHHHHHCCCCEEEEC | c1c-AT-4 | multi | 0.011 | 0.014 | 0.048 | ORD | 0.0 | |
| 23 | fp_O | Q99AM3 | 463 | LAAVLRSFASGLVIVS | HHHHHHHHCCCCEEEE | c1a-AT-4 | multi | 0.011 | 0.014 | 0.05 | ORD | 0.0 | |
| 24 | fp_O | Q99AM3 | 463 | LAAVLRSFASGLVIVS | HHHHHHHHCCCCEEEE | c1d-4 | multi | 0.011 | 0.014 | 0.05 | ORD | 0.0 | |
| 25 | fp_O | Q99AM3 | 466 | VLRSFASGLVIVSLRS | HHHHHCCCCEEEECCC | c1a-5 | multi-selected | 0.01 | 0.014 | 0.024 | ORD | 0.286 | |
| 26 | fp_O | Q99AM3 | 467 | LRSFASGLVIVSLRS | HHHHCCCCEEEECCC | c1b-AT-4 | multi | 0.01 | 0.013 | 0.023 | ORD | 0.429 | |
| 27 | fp_beta_O | Q99AM3 | 471 | ASGLVIVSLRSGIYVKN | CCCCEEEECCCEEEEEC | c1c-4 | multi | 0.01 | 0.015 | 0.024 | ORD | 0.625 | |
| 28 | fp_beta_O | Q99AM3 | 472 | SGLVIVSLRSGIYVKN | CCCEEEECCCEEEEEC | c1aR-4 | multi | 0.01 | 0.015 | 0.024 | ORD | 0.571 | |
| 29 | fp_beta_O | Q99AM3 | 472 | SGLVIVSLRSGIYVKN | CCCEEEECCCEEEEEC | c1d-4 | multi | 0.01 | 0.015 | 0.024 | ORD | 0.571 | |
| 30 | fp_O | Q99AM3 | 510 | LSSRAVLDVFNVAQ | CCHHHHHHHHHHHH | c2-AT-4 | uniq | 0.034 | 0.047 | 0.058 | ORD | 0.0 | |
| 31 | cand_beta_D | Q99AM3 | 547 | EQFDMVPLVIKLRLRS *+++++++++ | CCCCCCCEEEEEEEEE | c1a-4 | multi-selected | 0.062 | 0.054 | 0.087 | boundary | 0.571 | |
| 32 | cand_beta_D | Q99AM3 | 547 | EQFDMVPLVIKLRLRS *+++++++++ | CCCCCCCEEEEEEEEE | c1d-4 | multi | 0.062 | 0.054 | 0.087 | boundary | 0.571 | |
| 33 | cand_beta_D | Q99AM3 | 548 | QFDMVPLVIKLRLRS *+++++++++ | CCCCCCEEEEEEEEE | c1b-5 | multi | 0.058 | 0.048 | 0.087 | boundary | 0.714 | |
| 34 | cand_beta_D | Q99AM3 | 551 | MVPLVIKLRLRS *+++++++++ | CCCEEEEEEEEE | c2-rev | multi-selected | 0.043 | 0.039 | 0.076 | boundary | 1.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment