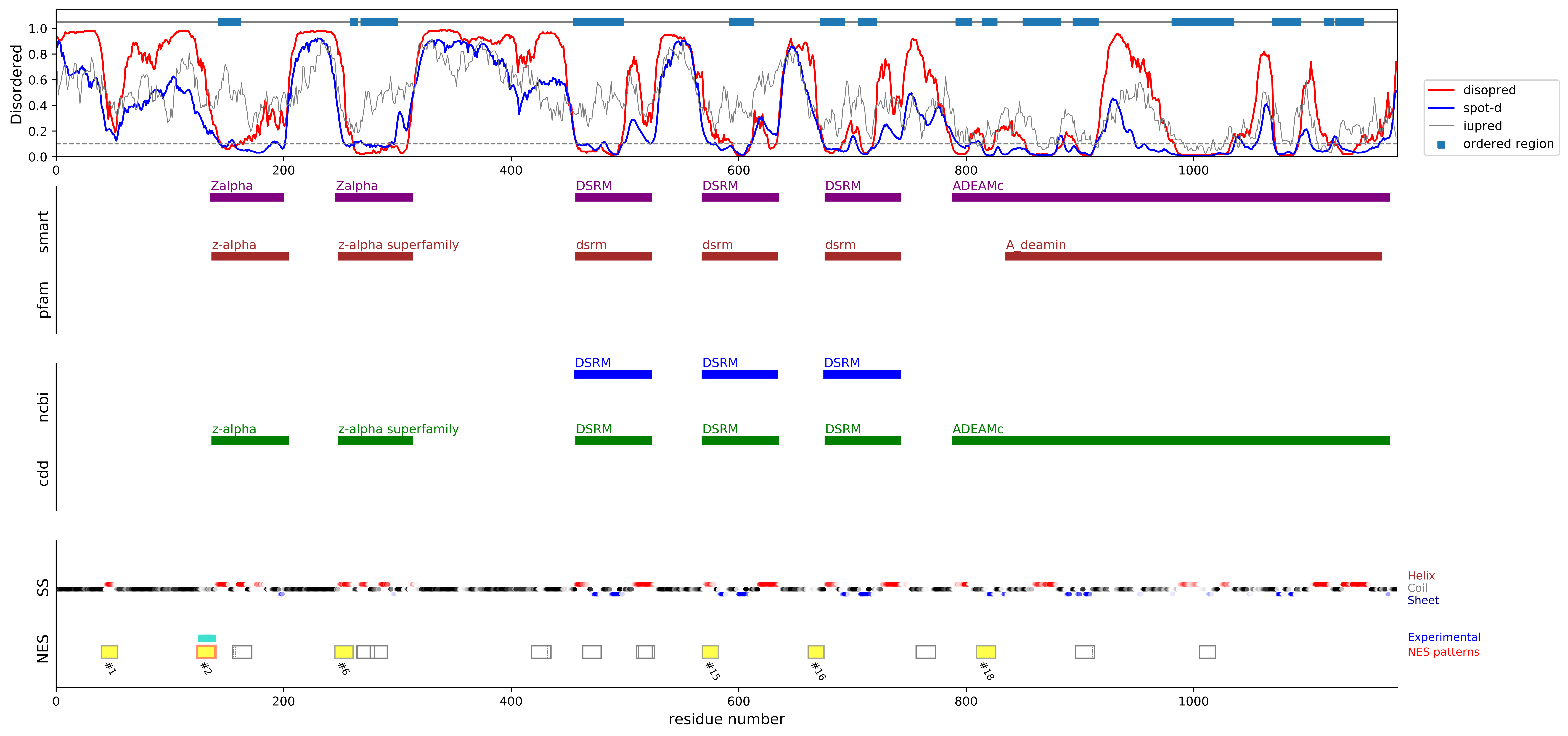

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q99MU3 | 40 | YPRSFQLQQIEFLK | CCCCHHHHHHHHHC | c2-4 | uniq | 0.416 | 0.227 | 0.452 | DISO | 0.0 | |
| 2 | cand_D | Q99MU3 | 124 | GADGLCSHFRELSISQ ++++++++++ | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.551 | 0.241 | 0.434 | boundary | boundary|z-alpha; | 0.0 |
| 3 | fp_D | Q99MU3 | 155 | GKATTAHVLARELRIPK | CCCCHHHHHHHHHCCCC | c1c-AT-4 | multi-selected | 0.112 | 0.064 | 0.432 | boundary | boundary|z-alpha; | 0.0 |
| 4 | fp_D | Q99MU3 | 156 | KATTAHVLARELRIPK | CCCHHHHHHHHHCCCC | c1d-AT-4 | multi | 0.113 | 0.06 | 0.431 | boundary | boundary|z-alpha; | 0.0 |
| 5 | fp_D | Q99MU3 | 158 | TTAHVLARELRIPK | CHHHHHHHHHCCCC | c2-AT-4 | multi | 0.118 | 0.054 | 0.42 | boundary | MID|z-alpha; | 0.0 |
| 6 | fp_D | Q99MU3 | 245 | EPLDMAEIKEKICDYL | CCCCHHHHHHHHHHHH | c1aR-4 | uniq | 0.549 | 0.209 | 0.388 | boundary | boundary|z-alpha superfamily; | 0.0 |
| 7 | fp_D | Q99MU3 | 264 | SNSSALNLAKNIGLTK | CCCHHHHHHHHHCCCC | c1d-AT-4 | multi-selected | 0.028 | 0.098 | 0.352 | boundary | boundary|z-alpha superfamily; | 0.0 |
| 8 | fp_D | Q99MU3 | 265 | NSSALNLAKNIGLTK | CCHHHHHHHHHCCCC | c1b-AT-4 | multi-selected | 0.028 | 0.098 | 0.357 | boundary | boundary|z-alpha superfamily; | 0.0 |
| 9 | fp_D | Q99MU3 | 276 | GLTKARDVTSVLIDL | CCCCCCCCCHHHHHH | c1b-AT-5 | uniq | 0.043 | 0.081 | 0.439 | boundary | MID|z-alpha superfamily; | 0.0 |
| 10 | fp_D | Q99MU3 | 418 | SIHTAPGEFRAIME | CCCCCCCCCCCCCC | c3-AT | multi | 0.901 | 0.562 | 0.512 | DISO | 0.0 | |
| 11 | fp_D | Q99MU3 | 418 | SIHTAPGEFRAIMEMPS | CCCCCCCCCCCCCCCCC | c1c-AT-5 | multi-selected | 0.912 | 0.564 | 0.492 | DISO | 0.0 | |
| 12 | fp_D | Q99MU3 | 463 | EYAQFTSQTCDFNLIE | HHHHHCCCEEEEEEEE | c1a-AT-4 | uniq | 0.045 | 0.077 | 0.387 | boundary | boundary|DSRM; | 0.429 |
| 13 | fp_D | Q99MU3 | 510 | AKQDAAVKAMAILL | HHHHHHHHHHHHHH | c2-AT-4 | multi-selected | 0.414 | 0.166 | 0.38 | boundary | boundary|DSRM; | 0.0 |
| 14 | fp_D | Q99MU3 | 512 | QDAAVKAMAILLRE | HHHHHHHHHHHHHH | c2-AT-4 | multi-selected | 0.366 | 0.147 | 0.37 | DISO | boundary|DSRM; | 0.0 |
| 15 | fp_D | Q99MU3 | 568 | PVTTLLECMHKLGN | CCHHHHHHHHHCCC | c3-4 | uniq | 0.293 | 0.128 | 0.322 | boundary | boundary|DSRM; | 0.0 |
| 16 | fp_D | Q99MU3 | 661 | KIRRIGELVRYLNT | CCCCCCCCCCCCCC | c3-4 | uniq | 0.506 | 0.294 | 0.322 | boundary | boundary|DSRM; boundary|DSRM; | 0.0 |
| 17 | fp_D | Q99MU3 | 756 | VTPVTGASLRRTMLLLS | CCCCCCCCCCHHHHCCC | c1c-AT-4 | uniq | 0.714 | 0.305 | 0.333 | DISO | boundary|DSRM; boundary|ADEAMc; | 0.0 |
| 18 | fp_D | Q99MU3 | 809 | QPSLLGRKILAAIIMKR | CCCCCCCCCEEEEEEEE | c1c-4 | uniq | 0.121 | 0.05 | 0.152 | boundary | MID|ADEAMc; | 0.375 |
| 19 | fp_beta_D | Q99MU3 | 896 | EKLQIKKTVSFHLYI | CCEEECCCEEEEEEC | c1b-AT-4 | multi | 0.031 | 0.032 | 0.148 | boundary | MID|ADEAMc; | 0.571 |
| 20 | fp_beta_D | Q99MU3 | 896 | EKLQIKKTVSFHLYIST | CCEEECCCEEEEEECCC | c1c-4 | multi-selected | 0.032 | 0.034 | 0.141 | boundary | MID|ADEAMc; | 0.625 |
| 21 | fp_beta_D | Q99MU3 | 1005 | FLQPVYLKSVTLGY | HCCCCEEEEEEECC | c2-5 | uniq | 0.012 | 0.008 | 0.073 | boundary | MID|ADEAMc; | 0.714 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment