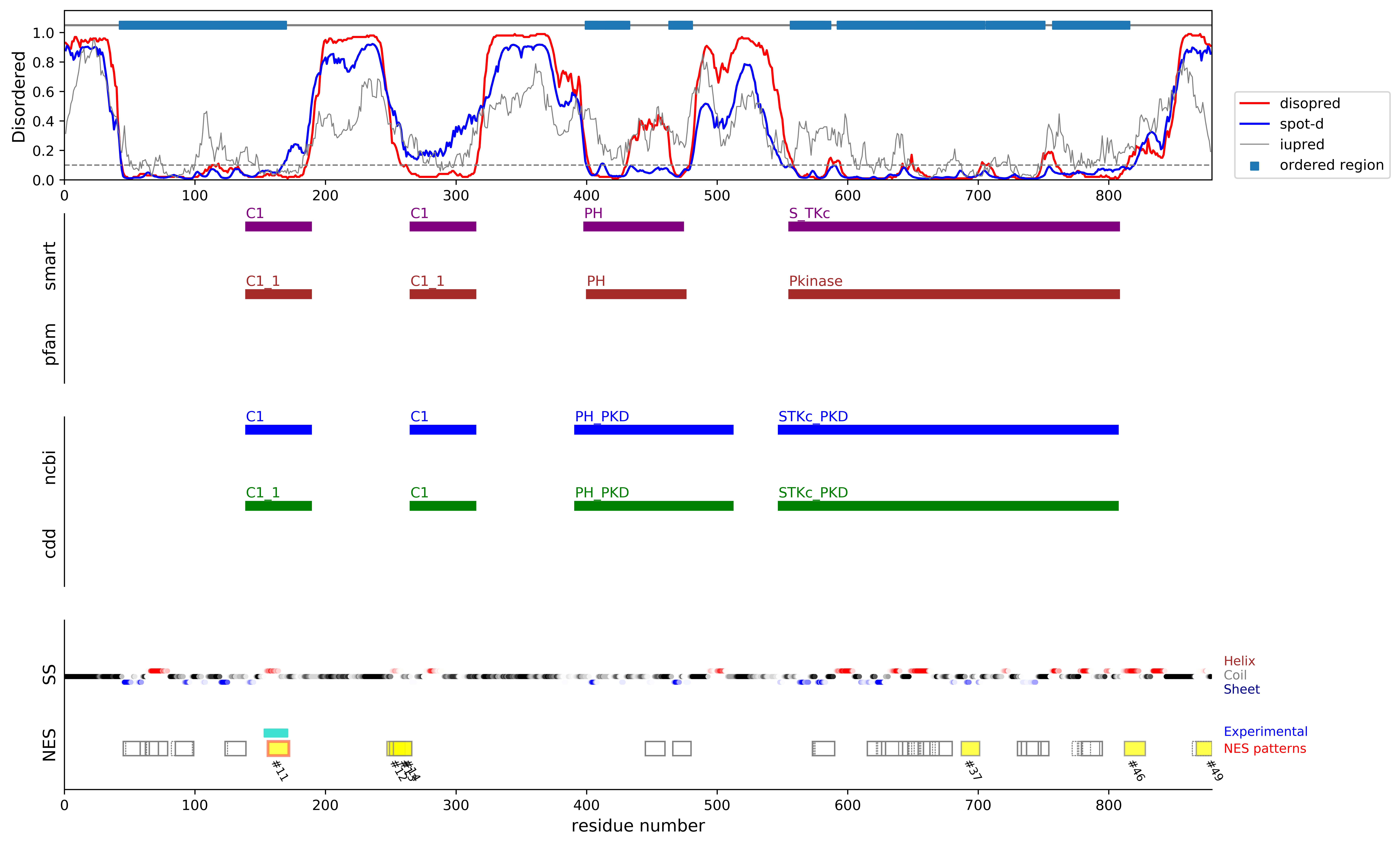

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | Q9BZL6 | 45 | VSFHIQIGLTREFVLLP | EEEEEEECCCEEEEECC | c1cR-5 | multi | 0.025 | 0.015 | 0.137 | boundary | 0.625 | |
| 2 | fp_beta_D | Q9BZL6 | 45 | VSFHIQIGLTREFVLLP | EEEEEEECCCEEEEECC | c1c-4 | multi-selected | 0.025 | 0.015 | 0.137 | boundary | 0.625 | |
| 3 | fp_beta_D | Q9BZL6 | 47 | FHIQIGLTREFVLLPA | EEEEECCCEEEEECCC | c1d-AT-4 | multi | 0.024 | 0.016 | 0.114 | boundary | 0.571 | |
| 4 | fp_beta_D | Q9BZL6 | 47 | FHIQIGLTREFVLLP | EEEEECCCEEEEECC | c1b-AT-4 | multi | 0.023 | 0.015 | 0.113 | boundary | 0.571 | |
| 5 | fp_D | Q9BZL6 | 58 | VLLPAASELAHVKQ | EECCCCHHHHHHHH | c3-AT | uniq | 0.033 | 0.03 | 0.101 | __ | 0.0 | |
| 6 | fp_O | Q9BZL6 | 65 | ELAHVKQLACSIVD | HHHHHHHHHHHHHH | c3-AT | uniq | 0.024 | 0.023 | 0.09 | ORD | 0.0 | |
| 7 | fp_O | Q9BZL6 | 82 | PECGFYGLYDKILLFK | CCCCCCCCCCEEEEEC | c1d-4 | multi | 0.017 | 0.018 | 0.041 | ORD | 0.143 | |
| 8 | fp_O | Q9BZL6 | 85 | GFYGLYDKILLFKH | CCCCCCCEEEEECC | c3-4 | multi-selected | 0.016 | 0.015 | 0.048 | ORD | 0.429 | |
| 9 | fp_O | Q9BZL6 | 123 | VVLSASATFEDFQIRP | EEECCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.065 | 0.043 | 0.149 | ORD | boundary|C1_1; | 0.0 |
| 10 | fp_O | Q9BZL6 | 125 | LSASATFEDFQIRP | ECCCCCCCCCCCCC | c2-AT-4 | multi | 0.066 | 0.047 | 0.144 | ORD | boundary|C1_1; | 0.0 |
| 11 | cand_D | Q9BZL6 | 156 | GEMLFGLVRQGLKCDG +++++++++++++ | HHHHHHHHHCCCCCCC | c1aR-4 | uniq | 0.029 | 0.079 | 0.053 | boundary | boundary|C1_1; boundary|C1_1; | 0.0 |
| 12 | fp_D | Q9BZL6 | 247 | TGRPIELDKMLLSK | CCCCHHHHHHHHCC | c2-4 | multi-selected | 0.265 | 0.422 | 0.351 | DISO | boundary|C1; | 0.0 |
| 13 | fp_D | Q9BZL6 | 249 | RPIELDKMLLSKVKVPH | CCHHHHHHHHCCCCCCC | c1c-4 | multi-selected | 0.174 | 0.333 | 0.315 | DISO | boundary|C1; | 0.0 |
| 14 | fp_D | Q9BZL6 | 252 | ELDKMLLSKVKVPH | HHHHHHHCCCCCCC | c2-5 | multi-selected | 0.132 | 0.298 | 0.304 | DISO | boundary|C1; | 0.0 |
| 15 | fp_D | Q9BZL6 | 445 | EILTVESAQNFSLVP | CCCEEECCCCCCCCC | c1b-AT-5 | uniq | 0.383 | 0.062 | 0.357 | boundary | MID|PH_PKD; | 0.286 |
| 16 | fp_D | Q9BZL6 | 466 | CFEIVTANATYFVG | EEEEEECCCCEEEC | c3-AT | uniq | 0.053 | 0.047 | 0.291 | boundary | MID|PH_PKD; | 0.429 |
| 17 | fp_beta_D | Q9BZL6 | 573 | TGRDVAVKVIDKLRFPT | CCCEEEEEEEECCCCCC | c1c-4 | multi-selected | 0.054 | 0.043 | 0.318 | boundary | MID|STKc_PKD; | 0.875 |
| 18 | fp_beta_D | Q9BZL6 | 574 | GRDVAVKVIDKLRFPT | CCEEEEEEEECCCCCC | c1a-AT-4 | multi | 0.056 | 0.042 | 0.32 | boundary | MID|STKc_PKD; | 0.857 |
| 19 | fp_beta_D | Q9BZL6 | 574 | GRDVAVKVIDKLRFPT | CCEEEEEEEECCCCCC | c1d-AT-4 | multi | 0.056 | 0.042 | 0.32 | boundary | MID|STKc_PKD; | 0.857 |
| 20 | fp_beta_D | Q9BZL6 | 575 | RDVAVKVIDKLRFPT | CEEEEEEEECCCCCC | c1b-4 | multi | 0.058 | 0.041 | 0.323 | boundary | MID|STKc_PKD; | 0.714 |
| 21 | fp_beta_O | Q9BZL6 | 615 | CMFETPEKVFVVME | EEEECCCEEEEEEE | c3-AT | uniq | 0.01 | 0.011 | 0.104 | ORD | MID|STKc_PKD; | 0.571 |
| 22 | fp_O | Q9BZL6 | 622 | KVFVVMEKLHGDMLEMI | EEEEEEECCCHHHHHHH | c1cR-5 | multi | 0.015 | 0.02 | 0.135 | ORD | MID|STKc_PKD; | 0.375 |
| 23 | fp_O | Q9BZL6 | 623 | VFVVMEKLHGDMLEMI | EEEEEECCCHHHHHHH | c1aR-5 | multi | 0.016 | 0.021 | 0.137 | ORD | MID|STKc_PKD; | 0.286 |
| 24 | fp_O | Q9BZL6 | 626 | VMEKLHGDMLEMILSS | EEECCCHHHHHHHHHC | c1a-5 | multi-selected | 0.024 | 0.028 | 0.167 | ORD | MID|STKc_PKD; | 0.0 |
| 25 | fp_O | Q9BZL6 | 642 | EKGRLPERLTKFLITQ | CCCCCCHHHHHHHHHH | c1a-4 | multi-selected | 0.083 | 0.035 | 0.103 | ORD | MID|STKc_PKD; | 0.0 |
| 26 | fp_O | Q9BZL6 | 646 | LPERLTKFLITQILVAL | CCHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.065 | 0.018 | 0.061 | ORD | MID|STKc_PKD; | 0.0 |
| 27 | fp_O | Q9BZL6 | 647 | PERLTKFLITQILVAL | CHHHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.065 | 0.015 | 0.056 | ORD | MID|STKc_PKD; | 0.0 |
| 28 | fp_O | Q9BZL6 | 647 | PERLTKFLITQILVAL | CHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.065 | 0.015 | 0.056 | ORD | MID|STKc_PKD; | 0.0 |
| 29 | fp_O | Q9BZL6 | 649 | RLTKFLITQILVAL | HHHHHHHHHHHHHH | c2-5 | multi | 0.06 | 0.013 | 0.047 | ORD | MID|STKc_PKD; | 0.0 |
| 30 | fp_O | Q9BZL6 | 649 | RLTKFLITQILVALRH | HHHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.054 | 0.013 | 0.043 | ORD | MID|STKc_PKD; | 0.0 |
| 31 | fp_O | Q9BZL6 | 651 | TKFLITQILVALRHLH | HHHHHHHHHHHHHHCC | c1aR-5 | multi | 0.039 | 0.012 | 0.032 | ORD | MID|STKc_PKD; | 0.0 |
| 32 | fp_O | Q9BZL6 | 654 | LITQILVALRHLHFKN | HHHHHHHHHHHCCCCC | c1a-5 | multi-selected | 0.029 | 0.012 | 0.029 | ORD | MID|STKc_PKD; | 0.0 |
| 33 | fp_O | Q9BZL6 | 654 | LITQILVALRHLHFKN | HHHHHHHHHHHCCCCC | c1d-AT-5 | multi | 0.029 | 0.012 | 0.029 | ORD | MID|STKc_PKD; | 0.0 |
| 34 | fp_O | Q9BZL6 | 655 | ITQILVALRHLHFKN | HHHHHHHHHHCCCCC | c1b-4 | multi | 0.025 | 0.012 | 0.027 | ORD | MID|STKc_PKD; | 0.0 |
| 35 | fp_O | Q9BZL6 | 656 | TQILVALRHLHFKN | HHHHHHHHHCCCCC | c2-4 | multi | 0.023 | 0.012 | 0.027 | ORD | MID|STKc_PKD; | 0.0 |
| 36 | fp_O | Q9BZL6 | 663 | RHLHFKNIVHCDLKPEN | HHCCCCCCCCCCCCCCC | c1cR-4 | uniq | 0.011 | 0.016 | 0.066 | ORD | MID|STKc_PKD; | 0.0 |
| 37 | fp_D | Q9BZL6 | 687 | PFPQVKLCDFGFAR | CCCCEEECCCCCCE | c2-5 | uniq | 0.016 | 0.029 | 0.045 | boundary | MID|STKc_PKD; | 0.429 |
| 38 | fp_beta_D | Q9BZL6 | 730 | RSLDMWSVGVIMYVSL | CCCCEEEEEEEEEEEC | c1d-4 | multi-selected | 0.011 | 0.011 | 0.034 | boundary | MID|STKc_PKD; | 1.0 |
| 39 | fp_beta_D | Q9BZL6 | 733 | DMWSVGVIMYVSLSG | CEEEEEEEEEEECCC | c1b-5 | multi-selected | 0.015 | 0.009 | 0.039 | boundary | MID|STKc_PKD; | 1.0 |
| 40 | fp_beta_D | Q9BZL6 | 737 | VGVIMYVSLSGTFPFNE | EEEEEEEECCCCCCCCC | c1cR-4 | multi-selected | 0.062 | 0.019 | 0.094 | boundary | MID|STKc_PKD; | 0.5 |
| 41 | fp_D | Q9BZL6 | 772 | PWSHISAGAIDLIN | CCCCCCHHHHHHHH | c2-AT-4 | multi | 0.039 | 0.046 | 0.122 | boundary | boundary|STKc_PKD; | 0.0 |
| 42 | fp_O | Q9BZL6 | 776 | ISAGAIDLINNLLQVKM | CCHHHHHHHHHHCCCCC | c1c-AT-4 | multi | 0.031 | 0.047 | 0.12 | ORD | boundary|STKc_PKD; | 0.0 |
| 43 | fp_O | Q9BZL6 | 777 | SAGAIDLINNLLQVKM | CHHHHHHHHHHCCCCC | c1d-4 | multi | 0.029 | 0.046 | 0.121 | ORD | boundary|STKc_PKD; | 0.0 |
| 44 | fp_O | Q9BZL6 | 779 | GAIDLINNLLQVKMRK | HHHHHHHHHCCCCCCC | c1a-4 | multi-selected | 0.025 | 0.049 | 0.126 | ORD | boundary|STKc_PKD; | 0.0 |
| 45 | fp_O | Q9BZL6 | 780 | AIDLINNLLQVKMRK | HHHHHHHHCCCCCCC | c1b-5 | multi | 0.023 | 0.049 | 0.122 | ORD | boundary|STKc_PKD; | 0.0 |
| 46 | fp_D | Q9BZL6 | 812 | TWLDLRELEGKMGERY | HHHHHHHHHHHHCCCC | c1aR-4 | uniq | 0.168 | 0.134 | 0.282 | boundary | boundary|STKc_PKD; | 0.0 |
| 47 | fp_D | Q9BZL6 | 864 | QDHDMQGLAERISVLX | CCCCCHHHHHHHCCC | c1a-AT-4 | multi | 0.959 | 0.873 | 0.448 | DISO | 0.0 | |
| 48 | fp_D | Q9BZL6 | 864 | QDHDMQGLAERISVLX | CCCCCHHHHHHHCCC | c1d-4 | multi | 0.959 | 0.873 | 0.448 | DISO | 0.0 | |
| 49 | fp_D | Q9BZL6 | 867 | DMQGLAERISVLXX | CCHHHHHHHCCC | c3-4-Ct | multi-selected | 0.953 | 0.868 | 0.406 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment