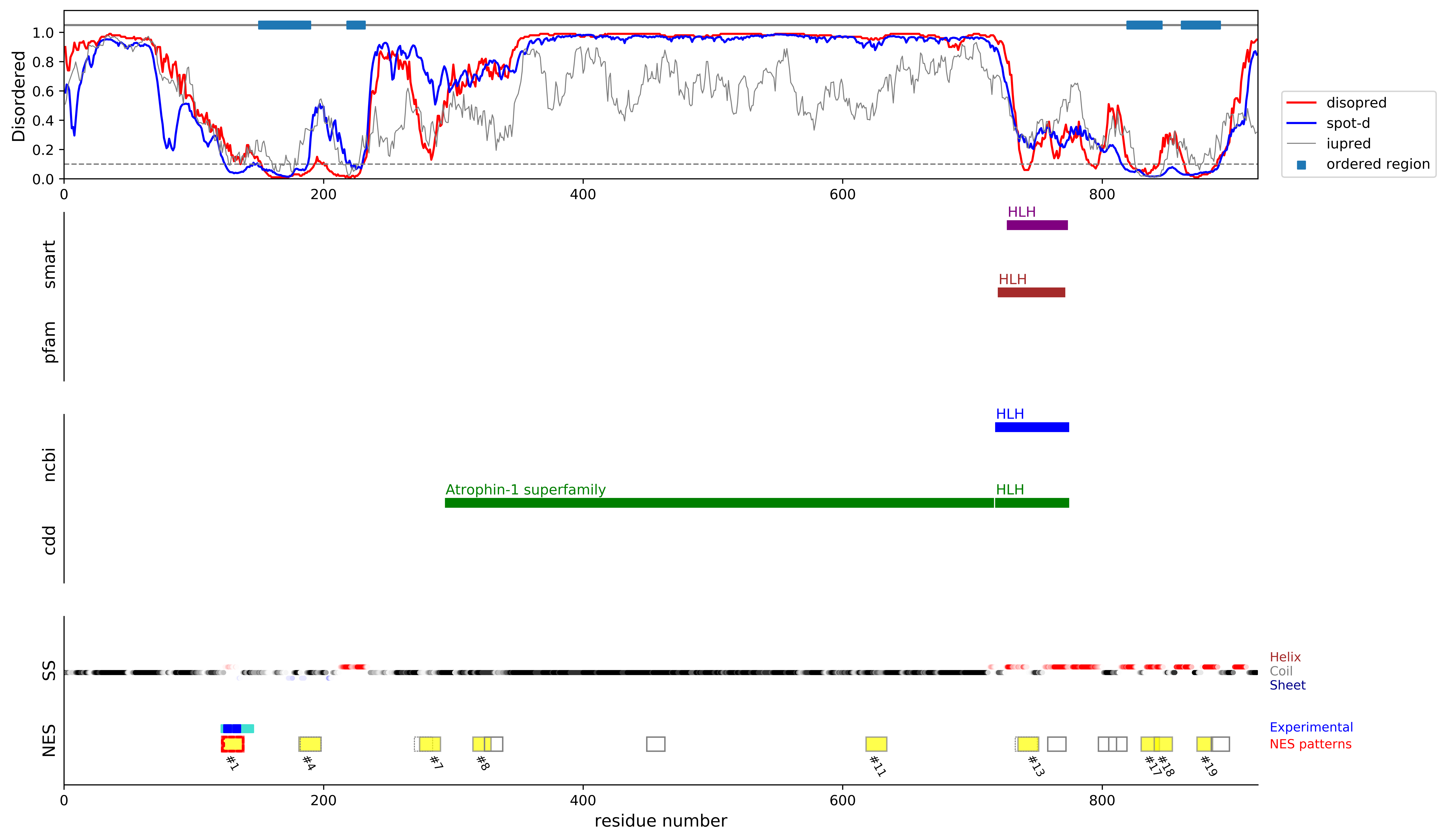

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | Q9HAP2 | 122 | IDASLTKLFECMTLAY ++*++++++*++++ | CCHHHHHHHHHHEEEC | c1a-4 | multi-selected | 0.181 | 0.061 | 0.155 | __ | 0.0 | |
| 2 | cand_D | Q9HAP2 | 122 | IDASLTKLFECMTLAY ++*++++++*++++ | CCHHHHHHHHHHEEEC | c1d-4 | multi | 0.181 | 0.061 | 0.155 | __ | 0.0 | |
| 3 | cand_D | Q9HAP2 | 123 | DASLTKLFECMTLAY ++*++++++*++++ | CHHHHHHHHHHEEEC | c1b-AT-4 | multi | 0.174 | 0.056 | 0.153 | __ | 0.0 | |
| 4 | fp_D | Q9HAP2 | 181 | PVCHFVTPLDGSVDVDE | CCEEECCCCCCCCCCCC | c1c-5 | multi-selected | 0.066 | 0.244 | 0.331 | boundary | 0.125 | |
| 5 | fp_D | Q9HAP2 | 182 | VCHFVTPLDGSVDVDE | CEEECCCCCCCCCCCC | c1d-4 | multi | 0.068 | 0.255 | 0.34 | boundary | 0.0 | |
| 6 | fp_D | Q9HAP2 | 270 | PLLDTDMLMSEFSD | CCCCCCCCCCCCCC | c3-AT | multi | 0.299 | 0.774 | 0.377 | DISO | boundary|Atrophin-1 superfamily; | 0.0 |
| 7 | fp_D | Q9HAP2 | 274 | TDMLMSEFSDTLFSTL | CCCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.256 | 0.68 | 0.33 | DISO | boundary|Atrophin-1 superfamily; | 0.0 |
| 8 | fp_D | Q9HAP2 | 315 | GLIPLQPNLDFMDT | CCCCCCCCCCCCCC | c3-4 | uniq | 0.746 | 0.729 | 0.405 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 9 | fp_D | Q9HAP2 | 324 | DFMDTFEPFQDLFS | CCCCCCCCCCCCCC | c3-AT | uniq | 0.744 | 0.774 | 0.333 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 10 | fp_D | Q9HAP2 | 449 | VLPLVPPPATALNP | CCCCCCCCCCCCCC | c3-AT | uniq | 0.988 | 0.97 | 0.721 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 11 | fp_D | Q9HAP2 | 618 | RAPGVPEFHSSILVTD | CCCCCCCCCCCCCCCC | c1d-4 | uniq | 0.958 | 0.922 | 0.527 | DISO | MID|Atrophin-1 superfamily; | 0.0 |
| 12 | fp_D | Q9HAP2 | 733 | FNIKMCFDMLNSLISNN | HHHHHHHHHHCCCCCCC | c1cR-4 | multi | 0.166 | 0.266 | 0.286 | DISO | boundary|Atrophin-1 superfamily; boundary|HLH; | 0.0 |
| 13 | fp_D | Q9HAP2 | 735 | IKMCFDMLNSLISNNS | HHHHHHHHCCCCCCCC | c1aR-4 | multi-selected | 0.143 | 0.258 | 0.284 | DISO | boundary|Atrophin-1 superfamily; boundary|HLH; | 0.0 |
| 14 | fp_D | Q9HAP2 | 758 | TLQKTVEYITKLQQ | HHHHHHHHHHHHHH | c3-AT | uniq | 0.244 | 0.275 | 0.407 | DISO | boundary|HLH; | 0.0 |
| 15 | fp_D | Q9HAP2 | 797 | CQQLLPATGVPVTR | HHHHCCCCCCCCCC | c2-AT-4 | uniq | 0.323 | 0.21 | 0.284 | boundary | boundary|HLH; | 0.0 |
| 16 | fp_D | Q9HAP2 | 805 | GVPVTRRQFDHMKD | CCCCCCCCCHHHHH | c3-AT | uniq | 0.394 | 0.162 | 0.346 | boundary | boundary|HLH; | 0.0 |
| 17 | fp_D | Q9HAP2 | 830 | QNWKFWIFSIIIKP | CCCCHHHHHHHHHH | c2-4 | uniq | 0.059 | 0.022 | 0.017 | boundary | 0.0 | |
| 18 | fp_D | Q9HAP2 | 840 | IIKPLFESFKGMVS | HHHHHHHHCCCCCC | c3-4 | uniq | 0.195 | 0.037 | 0.099 | boundary | 0.0 | |
| 19 | fp_D | Q9HAP2 | 873 | SLPILRPMVLST | CCCCHHHHHHHH | c2-rev | uniq | 0.028 | 0.042 | 0.131 | boundary | 0.0 | |
| 20 | fp_D | Q9HAP2 | 884 | TLRQLSTSTSILTD | HHHHHHCCCCCCCC | c3-AT | uniq | 0.14 | 0.123 | 0.281 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment