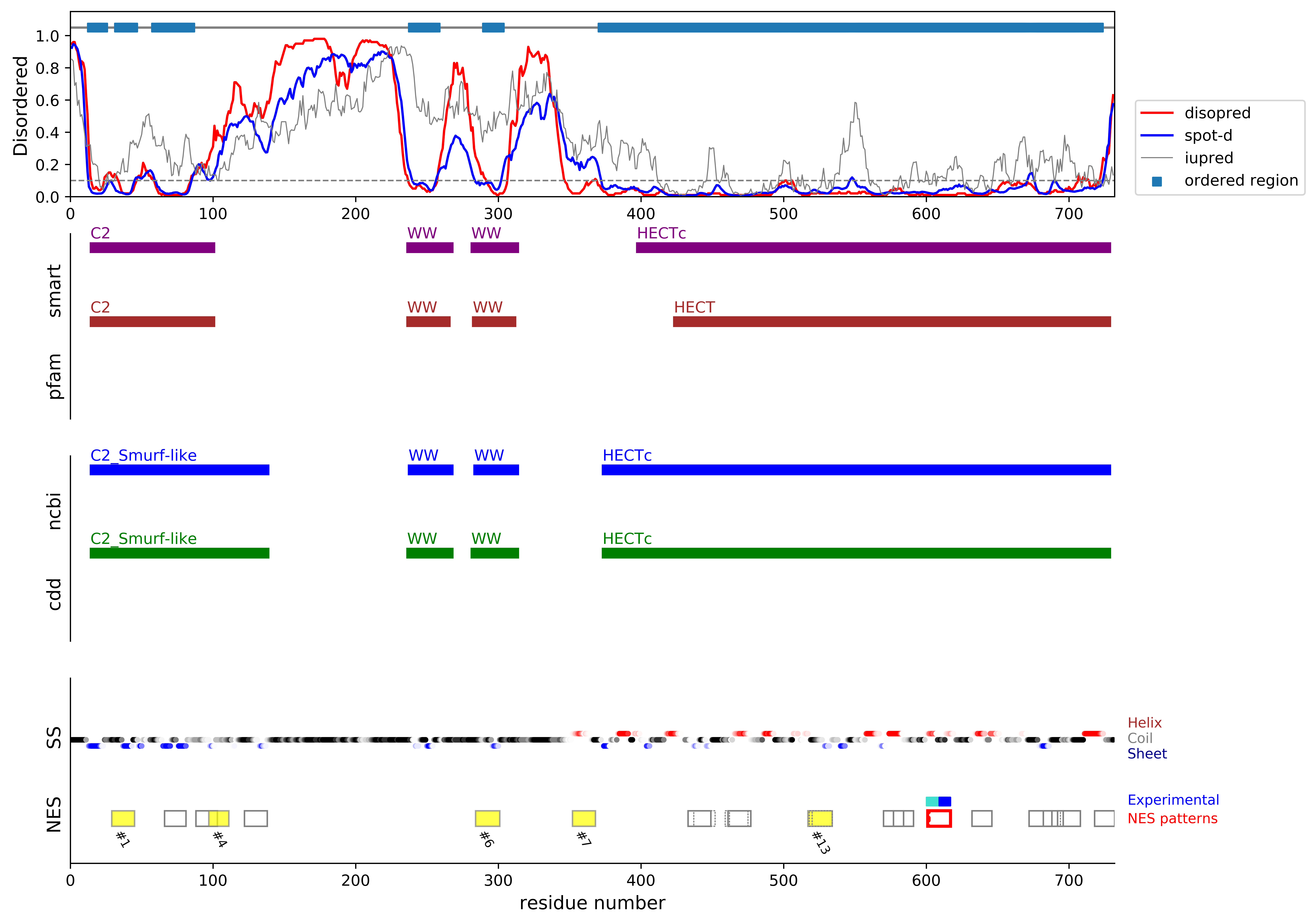

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9HCE7 | 29 | DFFRLPDPFAKIVVDG | CCCCCCCCEEEEEEEC | c1a-5 | uniq | 0.063 | 0.037 | 0.19 | boundary | boundary|C2_Smurf-like; | 0.429 |
| 2 | fp_D | Q9HCE7 | 66 | YDLYVGKTDSITISV | EEEEECCCCEEEEEE | c1b-AT-4 | uniq | 0.011 | 0.02 | 0.219 | boundary | MID|C2_Smurf-like; | 0.429 |
| 3 | fp_D | Q9HCE7 | 88 | KKQGAGFLGCVRLLS | CCCCCCCCCCEEEEC | c1b-AT-4 | uniq | 0.236 | 0.149 | 0.155 | boundary | MID|C2_Smurf-like; | 0.143 |
| 4 | fp_D | Q9HCE7 | 97 | CVRLLSNAISRLKD | CEEEECCCCCCCCC | c3-4 | uniq | 0.368 | 0.228 | 0.153 | boundary | MID|C2_Smurf-like; | 0.286 |
| 5 | fp_D | Q9HCE7 | 122 | NPSDTDAVRGQIVVSL | CCCCCCCCCCEEEEEE | c1d-AT-4 | uniq | 0.529 | 0.406 | 0.409 | DISO | boundary|C2_Smurf-like; | 0.143 |
| 6 | fp_D | Q9HCE7 | 284 | PGWEVRSTVSGRIYFVD | CCCCCCCCCCCCEEEEE | c1c-4 | uniq | 0.102 | 0.075 | 0.45 | boundary | small|WW; | 0.0 |
| 7 | fp_D | Q9HCE7 | 352 | LVQKLKVLRHELSLQQ | HHHHHHHHHHHHHCCC | c1d-5 | uniq | 0.052 | 0.213 | 0.311 | boundary | boundary|HECTc; | 0.0 |
| 8 | fp_beta_O | Q9HCE7 | 433 | YYGLFQYSTDNIYMLQ | CCCCEEEECCCCCEEE | c1a-AT-4 | multi-selected | 0.011 | 0.014 | 0.079 | ORD | MID|HECTc; | 0.571 |
| 9 | fp_O | Q9HCE7 | 437 | FQYSTDNIYMLQINP | EEEECCCCCEEEECC | c1b-AT-4 | multi | 0.013 | 0.018 | 0.133 | ORD | MID|HECTc; | 0.286 |
| 10 | fp_O | Q9HCE7 | 459 | HLSYFHFVGRIMGLAV | CCCCCCHHHHHHHHHH | c1d-5 | multi | 0.01 | 0.01 | 0.046 | ORD | MID|HECTc; | 0.0 |
| 11 | fp_O | Q9HCE7 | 461 | SYFHFVGRIMGLAVFH | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.01 | 0.009 | 0.032 | ORD | MID|HECTc; | 0.0 |
| 12 | fp_O | Q9HCE7 | 462 | YFHFVGRIMGLAVFH | CCCHHHHHHHHHHHH | c1b-5 | multi | 0.01 | 0.008 | 0.026 | ORD | MID|HECTc; | 0.0 |
| 13 | fp_D | Q9HCE7 | 517 | LENDITPVLDHTFCVEH | HCCCCCCCCCCEEEEEC | c1c-4 | multi-selected | 0.019 | 0.029 | 0.064 | boundary | MID|HECTc; | 0.125 |
| 14 | fp_D | Q9HCE7 | 518 | ENDITPVLDHTFCVEH | CCCCCCCCCCEEEEEC | c1d-AT-4 | multi | 0.02 | 0.029 | 0.063 | boundary | MID|HECTc; | 0.143 |
| 15 | fp_D | Q9HCE7 | 520 | DITPVLDHTFCVEH | CCCCCCCCEEEEEC | c3-AT | multi | 0.021 | 0.03 | 0.066 | boundary | MID|HECTc; | 0.286 |
| 16 | fp_O | Q9HCE7 | 570 | FMRGIEAQFLALQK | EECCHHHHHHHHHH | c2-AT-5 | multi | 0.034 | 0.022 | 0.047 | ORD | MID|HECTc; | 0.0 |
| 17 | fp_O | Q9HCE7 | 570 | FMRGIEAQFLALQK | EECCHHHHHHHHHH | c3-4 | multi-selected | 0.034 | 0.022 | 0.047 | ORD | MID|HECTc; | 0.0 |
| 18 | fp_O | Q9HCE7 | 577 | QFLALQKGFNELIP | HHHHHHHCCCCCCC | c3-4 | uniq | 0.031 | 0.023 | 0.081 | ORD | MID|HECTc; | 0.0 |
| 19 | cand_O | Q9HCE7 | 601 | ELELIIGGLDKIDLND +++++++++*+* | HHHHHHHCCCCCCHHH | c1a-5 | multi-selected | 0.011 | 0.033 | 0.118 | ORD | MID|HECTc; | 0.0 |
| 20 | cand_O | Q9HCE7 | 602 | LELIIGGLDKIDLND +++++++++*+* | HHHHHHCCCCCCHHH | c1b-4 | multi | 0.011 | 0.033 | 0.117 | ORD | MID|HECTc; | 0.0 |
| 21 | fp_O | Q9HCE7 | 632 | IVRWFWQAVETFDE | HHHHHHHHHHCCCH | c3-4 | uniq | 0.011 | 0.025 | 0.071 | ORD | MID|HECTc; | 0.0 |
| 22 | fp_beta_D | Q9HCE7 | 672 | STGAAGPRLFTIHLID | CCCCCCCEEEEEEEEC | c1a-AT-4 | uniq | 0.038 | 0.059 | 0.211 | boundary | MID|HECTc; | 0.571 |
| 23 | fp_D | Q9HCE7 | 682 | TIHLIDANTDNLPK | EEEEECCCCCCCCC | c3-AT | uniq | 0.028 | 0.053 | 0.202 | boundary | MID|HECTc; | 0.286 |
| 24 | fp_D | Q9HCE7 | 692 | NLPKAHTCFNRIDIPP | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.052 | 0.041 | 0.224 | boundary | boundary|HECTc; | 0.0 |
| 25 | fp_D | Q9HCE7 | 694 | PKAHTCFNRIDIPP | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.056 | 0.037 | 0.229 | boundary | boundary|HECTc; | 0.0 |
| 26 | fp_D | Q9HCE7 | 718 | LLTAVEETCGFAVEX | HHHHHHHCCCCCCC | c1b-AT-5 | uniq | 0.244 | 0.219 | 0.102 | boundary | boundary|HECTc; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment