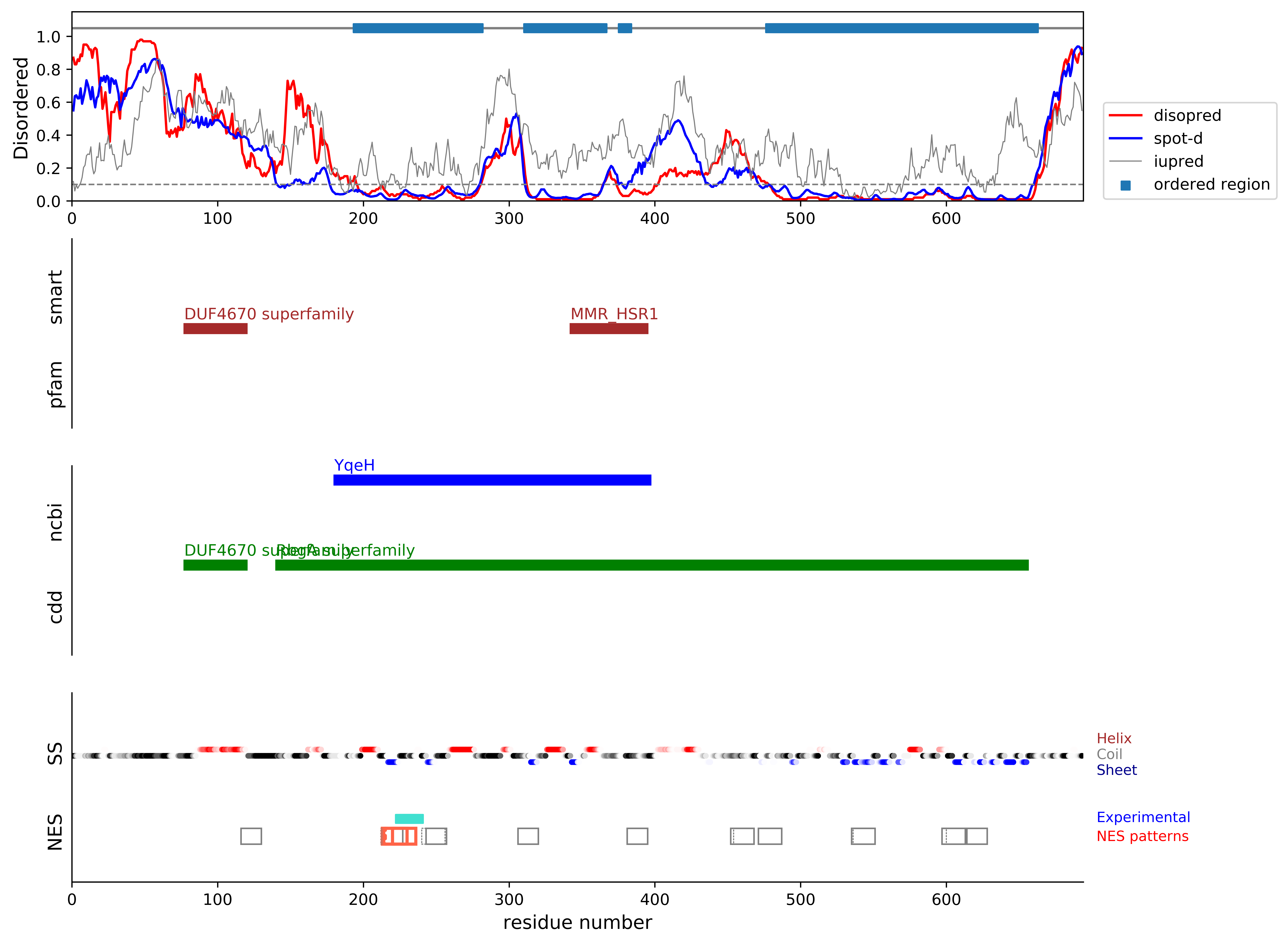

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9JJG9 | 116 | QKLRAGFRTLPVPE | HHHHCCCCCCCCCC | c2-AT-4 | uniq | 0.246 | 0.347 | 0.45 | DISO | boundary|RbgA superfamily; | 0.0 |
| 2 | fp_beta_O | Q9JJG9 | 212 | RPGPALVLYMVNLLD ++ | CCCCCEEEEEEEECC | c1b-AT-4 | multi | 0.049 | 0.019 | 0.115 | ORD | MID|RbgA superfamily; | 0.857 |
| 3 | cand_beta_O | Q9JJG9 | 213 | PGPALVLYMVNLLDLPD +++++ | CCCCEEEEEEEECCCCC | c1c-4 | multi-selected | 0.045 | 0.025 | 0.112 | ORD | MID|RbgA superfamily; | 1.0 |
| 4 | fp_beta_O | Q9JJG9 | 213 | PGPALVLYMVNLLD ++ | CCCCEEEEEEEECC | c2-4 | multi-selected | 0.046 | 0.017 | 0.118 | ORD | MID|RbgA superfamily; | 0.857 |
| 5 | cand_beta_O | Q9JJG9 | 214 | GPALVLYMVNLLDLPD +++++ | CCCEEEEEEEECCCCC | c1a-4 | multi-selected | 0.043 | 0.024 | 0.112 | ORD | MID|RbgA superfamily; | 1.0 |
| 6 | cand_beta_O | Q9JJG9 | 214 | GPALVLYMVNLLDLPD +++++ | CCCEEEEEEEECCCCC | c1d-4 | multi | 0.043 | 0.024 | 0.112 | ORD | MID|RbgA superfamily; | 1.0 |
| 7 | cand_beta_O | Q9JJG9 | 215 | PALVLYMVNLLDLPD +++++ | CCEEEEEEEECCCCC | c1b-4 | multi | 0.041 | 0.024 | 0.111 | ORD | MID|RbgA superfamily; | 0.857 |
| 8 | cand_O | Q9JJG9 | 220 | YMVNLLDLPDALLPDL +++++++++++ | EEEEECCCCCCCCCCC | c1aR-4 | multi-selected | 0.035 | 0.036 | 0.153 | ORD | MID|RbgA superfamily; | 0.143 |
| 9 | fp_beta_O | Q9JJG9 | 240 | GPKQLIVLGNKVDLLP | CCCCEEEEEECCCCCC | c1d-4 | multi | 0.045 | 0.027 | 0.206 | ORD | MID|RbgA superfamily; | 0.857 |
| 10 | fp_beta_O | Q9JJG9 | 243 | QLIVLGNKVDLLPQ | CEEEEEECCCCCCC | c3-4 | multi-selected | 0.052 | 0.029 | 0.211 | ORD | MID|RbgA superfamily; | 0.571 |
| 11 | fp_D | Q9JJG9 | 306 | SRSRTVVKDVRLIS | CCCCCCCCCEEEEE | c2-AT-4 | uniq | 0.152 | 0.15 | 0.397 | boundary | MID|RbgA superfamily; | 0.143 |
| 12 | fp_D | Q9JJG9 | 381 | PWPGTTLNLLKFPI | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.049 | 0.175 | 0.299 | boundary | MID|RbgA superfamily; | 0.0 |
| 13 | fp_D | Q9JJG9 | 452 | DEVPFEFDADSLAFDM | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.289 | 0.179 | 0.277 | boundary | MID|RbgA superfamily; | 0.0 |
| 14 | fp_D | Q9JJG9 | 454 | VPFEFDADSLAFDM | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.279 | 0.178 | 0.267 | boundary | MID|RbgA superfamily; | 0.0 |
| 15 | fp_D | Q9JJG9 | 471 | PVVSVCKSTKQIELTP | CCEEECCCCCEECCCC | c1a-AT-5 | uniq | 0.086 | 0.08 | 0.31 | boundary | MID|RbgA superfamily; | 0.286 |
| 16 | fp_beta_O | Q9JJG9 | 535 | GMVLFLGGIARIDFLQ | CCEEEEEEEEEEEEEC | c1a-5 | multi-selected | 0.016 | 0.009 | 0.039 | ORD | MID|RbgA superfamily; | 1.0 |
| 17 | fp_beta_O | Q9JJG9 | 536 | MVLFLGGIARIDFLQ | CEEEEEEEEEEEEEC | c1b-5 | multi | 0.016 | 0.009 | 0.039 | ORD | MID|RbgA superfamily; | 1.0 |
| 18 | fp_beta_O | Q9JJG9 | 597 | RMAQFPPLVAEDITLKG | HHHHCCCCEEEEEEEEC | c1c-5 | multi-selected | 0.022 | 0.032 | 0.26 | ORD | MID|RbgA superfamily; | 0.5 |
| 19 | fp_beta_O | Q9JJG9 | 600 | QFPPLVAEDITLKG | HCCCCEEEEEEEEC | c2-AT-5 | multi | 0.017 | 0.028 | 0.267 | ORD | MID|RbgA superfamily; | 0.714 |
| 20 | fp_beta_O | Q9JJG9 | 613 | GGGKFEAVADIKFSS | CCCCCEEEEEEEEEC | c1b-4 | uniq | 0.017 | 0.035 | 0.121 | ORD | MID|RbgA superfamily; | 0.857 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment