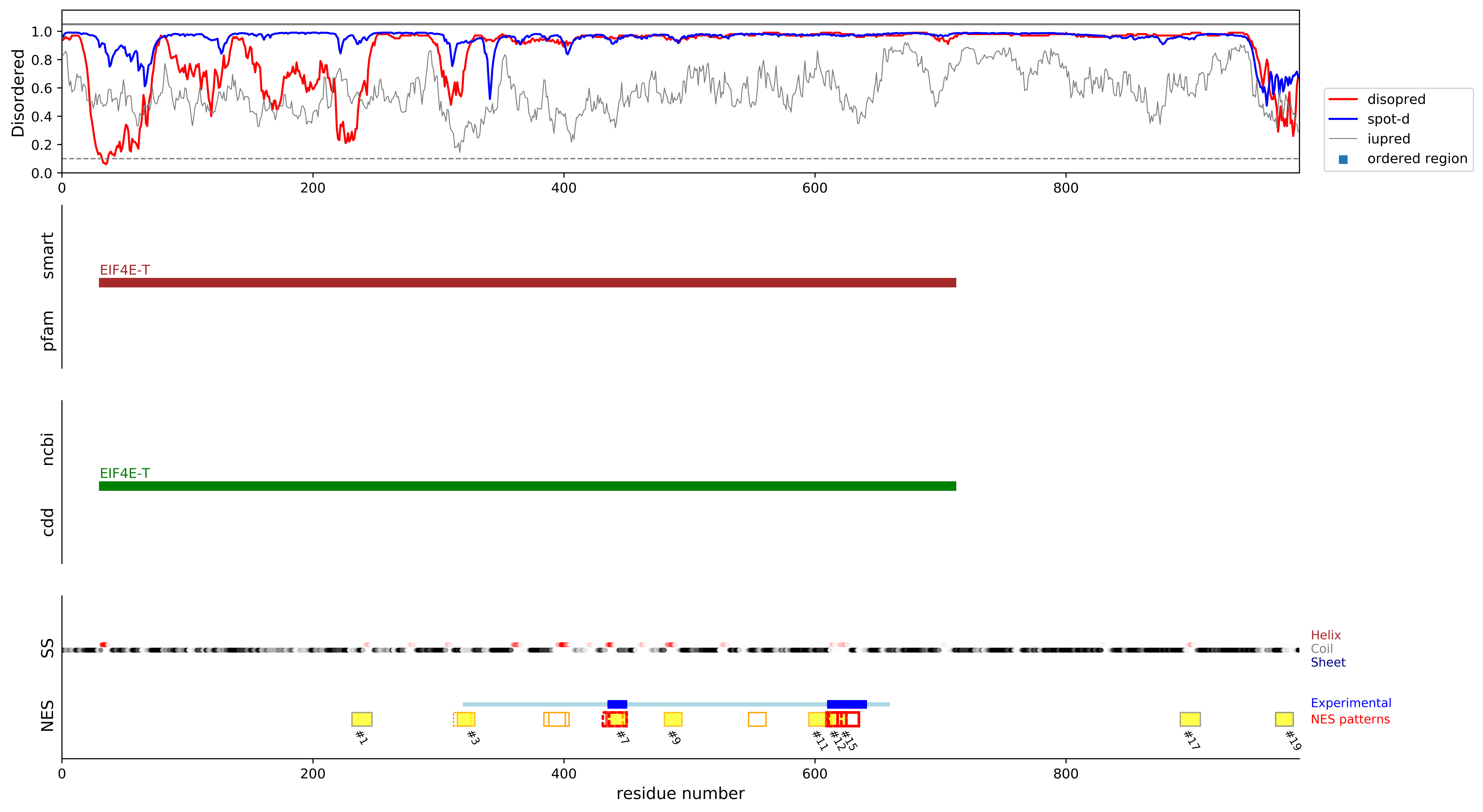

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9NRA8 | 231 | ETIELTGFDDKILEED | CCCCCCCCCCHHHHHH | c1aR-4 | uniq | 0.521 | 0.946 | 0.511 | DISO | MID|EIF4E-T; | 0.0 |
| 2 | fp_D | Q9NRA8 | 312 | NLDKVPCLASMIED ..... | CCCCCCCCCCCCCC | c3-AT | multi | 0.681 | 0.902 | 0.251 | DISO | MID|EIF4E-T; | 0.0 |
| 3 | fp_D | Q9NRA8 | 315 | KVPCLASMIEDVLG ........ | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.756 | 0.922 | 0.274 | DISO | MID|EIF4E-T; | 0.0 |
| 4 | fp_D | Q9NRA8 | 384 | APIPLEDHAENKVDILE ................. | CCCCCCCCCCHHHHHHH | c1c-AT-4 | multi-selected | 0.928 | 0.956 | 0.446 | DISO | MID|EIF4E-T; | 0.0 |
| 5 | fp_D | Q9NRA8 | 388 | LEDHAENKVDILEMLQ ................ | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.922 | 0.939 | 0.392 | DISO | MID|EIF4E-T; | 0.0 |
| 6 | cand_D | Q9NRA8 | 431 | VVLSVEEVEAGLKGLK .......*...*..*. | CCCHHHHHHHHHCCCC | c1aR-5 | multi | 0.957 | 0.942 | 0.435 | DISO | MID|EIF4E-T; | 0.0 |
| 7 | cand_D | Q9NRA8 | 434 | SVEEVEAGLKGLKVDQ ....*...*..*.*.. | HHHHHHHHHCCCCCCC | c1a-5 | multi-selected | 0.958 | 0.942 | 0.443 | DISO | MID|EIF4E-T; | 0.0 |
| 8 | cand_D | Q9NRA8 | 436 | EEVEAGLKGLKVDQ ..*...*..*.*.. | HHHHHHHCCCCCCC | c2-AT-4 | multi | 0.959 | 0.941 | 0.44 | DISO | MID|EIF4E-T; | 0.0 |
| 9 | fp_D | Q9NRA8 | 480 | DMTAFNKLVSTMKA .............. | CHHHHHHHHHCCCC | c3-4 | uniq | 0.942 | 0.952 | 0.529 | DISO | MID|EIF4E-T; | 0.0 |
| 10 | fp_D | Q9NRA8 | 547 | LMGSLEPTTSLLGQ .............. | CCCCCCCCCCCCCC | c3-AT | uniq | 0.977 | 0.98 | 0.605 | DISO | MID|EIF4E-T; | 0.0 |
| 11 | fp_D | Q9NRA8 | 595 | PQQLLGDPFQGMRKPMSP .................. | CCCCCCCCCCCCCCCCCC | c4-4 | uniq | 0.983 | 0.975 | 0.701 | DISO | MID|EIF4E-T; | 0.0 |
| 12 | cand_D | Q9NRA8 | 609 | PMSPITAQMSQLELQQ ....*...*..*.*.. | CCCCHHHHCCHHHHHH | c1a-5 | multi-selected | 0.987 | 0.972 | 0.604 | DISO | MID|EIF4E-T; | 0.0 |
| 13 | cand_D | Q9NRA8 | 610 | MSPITAQMSQLELQQ ...*...*..*.*.. | CCCHHHHCCHHHHHH | c1b-AT-4 | multi | 0.987 | 0.971 | 0.596 | DISO | MID|EIF4E-T; | 0.0 |
| 14 | cand_D | Q9NRA8 | 611 | SPITAQMSQLELQQ ..*...*..*.*.. | CCHHHHCCHHHHHH | c2-AT-4 | multi | 0.986 | 0.971 | 0.588 | DISO | MID|EIF4E-T; | 0.0 |
| 15 | cand_D | Q9NRA8 | 618 | SQLELQQAALEGLALPH ..*.*....*..*.*.. | CHHHHHHHHCCCCCCCC | c1c-AT-4 | multi-selected | 0.978 | 0.971 | 0.487 | DISO | MID|EIF4E-T; | 0.0 |
| 16 | cand_D | Q9NRA8 | 621 | ELQQAALEGLALPH .*....*..*.*.. | HHHHHHCCCCCCCC | c2-AT-5 | multi | 0.976 | 0.971 | 0.469 | DISO | MID|EIF4E-T; | 0.0 |
| 17 | fp_D | Q9NRA8 | 891 | TPLHLAMVQQQLQRSV | CCCCCCHHHHHHCCCC | c1aR-4 | uniq | 0.983 | 0.956 | 0.604 | DISO | 0.0 | |
| 18 | fp_D | Q9NRA8 | 967 | PLPSMPAKVISVDE | CCCCCCCCCCCHHC | c2-AT-5 | multi | 0.406 | 0.643 | 0.467 | DISO | 0.0 | |
| 19 | fp_D | Q9NRA8 | 967 | PLPSMPAKVISVDE | CCCCCCCCCCCHHC | c3-4 | multi-selected | 0.406 | 0.643 | 0.467 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment