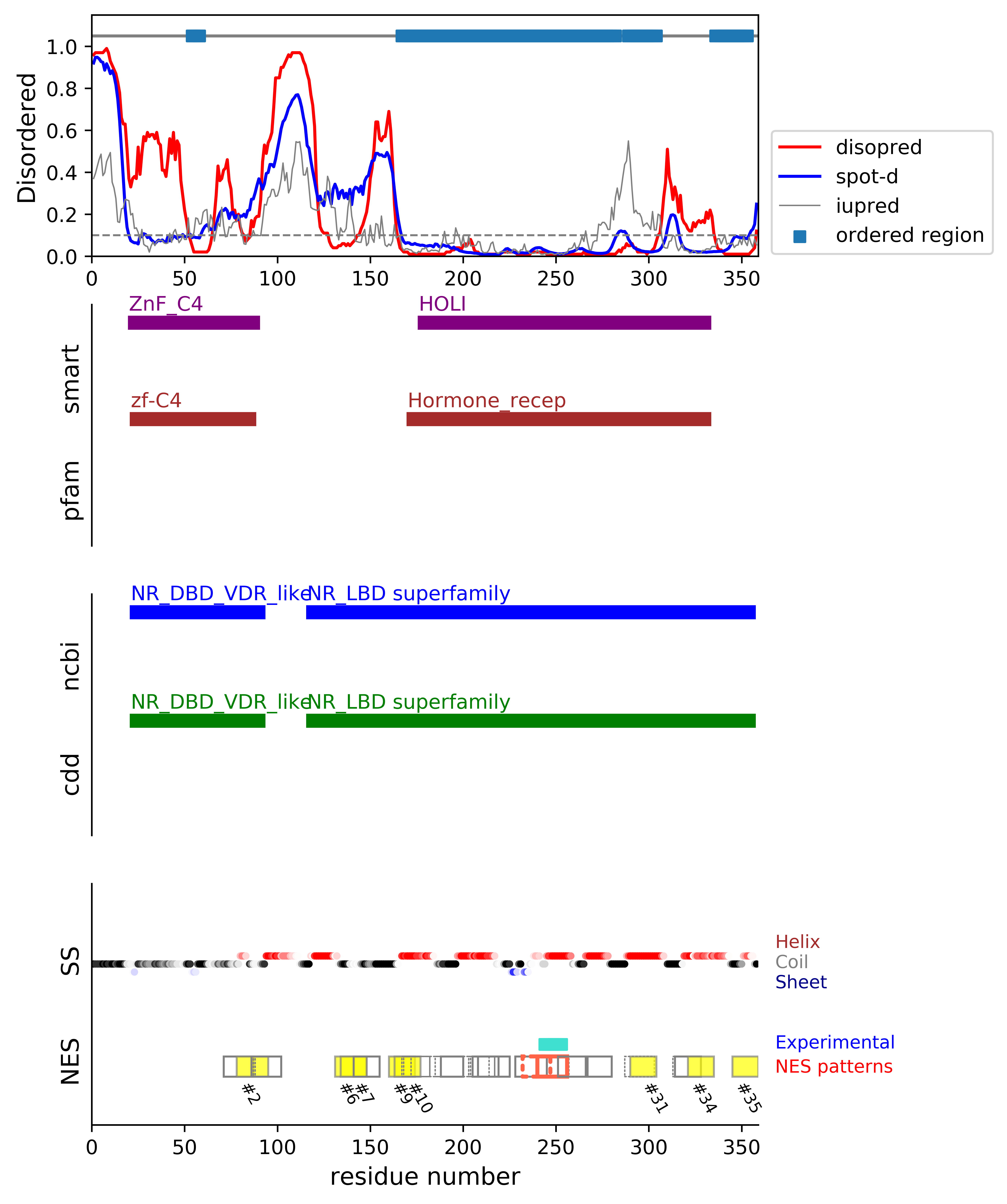

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9QUS1 | 71 | RHCPACRLQKCLNVGM | CCCCCCCCCHHHHCCC | c1d-AT-4 | uniq | 0.22 | 0.205 | 0.123 | DISO | boundary|NR_DBD_VDR_like; | 0.0 |
| 2 | fp_D | Q9QUS1 | 78 | LQKCLNVGMRKDMILSA | CCHHHHCCCCCCCCCCH | c1c-4 | uniq | 0.21 | 0.267 | 0.13 | DISO | boundary|NR_DBD_VDR_like; | 0.0 |
| 3 | fp_D | Q9QUS1 | 86 | MRKDMILSAEALALRR | CCCCCCCCHHHHHHHH | c1a-AT-4 | multi-selected | 0.472 | 0.387 | 0.21 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD superfamily; | 0.0 |
| 4 | fp_D | Q9QUS1 | 87 | RKDMILSAEALALRR | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.493 | 0.394 | 0.218 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD superfamily; | 0.0 |
| 5 | fp_D | Q9QUS1 | 88 | KDMILSAEALALRR | CCCCCCHHHHHHHH | c2-AT-4 | multi | 0.518 | 0.403 | 0.224 | DISO | boundary|NR_DBD_VDR_like; boundary|NR_LBD superfamily; | 0.0 |
| 6 | fp_D | Q9QUS1 | 131 | HTRHVGPMFDQFVQFRP | HHCCCCCCCCCCCCCCC | c1c-4 | multi-selected | 0.078 | 0.308 | 0.135 | DISO | boundary|NR_LBD superfamily; | 0.0 |
| 7 | fp_D | Q9QUS1 | 134 | HVGPMFDQFVQFRP | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.086 | 0.307 | 0.133 | DISO | boundary|NR_LBD superfamily; | 0.0 |
| 8 | fp_D | Q9QUS1 | 141 | QFVQFRPPAYLFSH | CCCCCCCCCCCCCC | c3-AT | uniq | 0.276 | 0.381 | 0.098 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 9 | fp_D | Q9QUS1 | 160 | PLAPVLPLLTHFAD | CCCCCCHHHHHHHH | c3-4 | multi-selected | 0.168 | 0.188 | 0.059 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 10 | fp_D | Q9QUS1 | 163 | PVLPLLTHFADINT | CCCHHHHHHHHHHH | c3-4 | multi-selected | 0.046 | 0.107 | 0.05 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 11 | fp_D | Q9QUS1 | 167 | LLTHFADINTFMVQQ | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.013 | 0.059 | 0.029 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 12 | fp_D | Q9QUS1 | 168 | LTHFADINTFMVQQ | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.012 | 0.057 | 0.028 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 13 | fp_D | Q9QUS1 | 172 | ADINTFMVQQIIKFTK | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.01 | 0.053 | 0.026 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 14 | fp_O | Q9QUS1 | 185 | FTKDLPLFRSLTMED | CCCCCCCCCCCCHHH | c1b-4 | multi | 0.022 | 0.043 | 0.066 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 15 | fp_O | Q9QUS1 | 188 | DLPLFRSLTMED | CCCCCCCCCHHH | c2-rev | multi-selected | 0.025 | 0.04 | 0.077 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 16 | fp_O | Q9QUS1 | 200 | QISLLKGAAVEILHISL | HHHHHHHHHHHHHHHHH | c1c-AT-5 | multi | 0.031 | 0.013 | 0.051 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 17 | fp_O | Q9QUS1 | 200 | QISLLKGAAVEILH | HHHHHHHHHHHHHH | c3-AT | multi | 0.035 | 0.014 | 0.05 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 18 | fp_O | Q9QUS1 | 203 | LLKGAAVEILHISL | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.026 | 0.012 | 0.048 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 19 | fp_O | Q9QUS1 | 203 | LLKGAAVEILHISLNT | HHHHHHHHHHHHHHHC | c1a-AT-5 | multi | 0.024 | 0.011 | 0.045 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 20 | fp_O | Q9QUS1 | 204 | LKGAAVEILHISLNT | HHHHHHHHHHHHHHC | c1b-AT-4 | multi | 0.021 | 0.011 | 0.042 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 21 | fp_O | Q9QUS1 | 205 | KGAAVEILHISLNT | HHHHHHHHHHHHHC | c2-4 | multi-selected | 0.016 | 0.011 | 0.041 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 22 | fp_O | Q9QUS1 | 208 | AVEILHISLNTTFCLQT | HHHHHHHHHHCCCCCCC | c1c-5 | multi-selected | 0.013 | 0.014 | 0.032 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 23 | fp_beta_O | Q9QUS1 | 228 | FCGPLCYKMEDAVHVGF + | EECCEEEECCHHHHCCC | c1c-4 | multi-selected | 0.015 | 0.025 | 0.022 | ORD | MID|NR_LBD superfamily; | 0.5 |
| 24 | cand_O | Q9QUS1 | 232 | LCYKMEDAVHVGFQY +++ | EEEECCHHHHCCCHH | c1b-AT-4 | multi | 0.015 | 0.028 | 0.02 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 25 | fp_O | Q9QUS1 | 240 | VHVGFQYEFLELIIHFH ++++++++++ | HHCCCHHHHHHHHHHHH | c1cR-5 | multi | 0.013 | 0.021 | 0.013 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 26 | cand_O | Q9QUS1 | 240 | VHVGFQYEFLELIIHF ++++++++++ | HHCCCHHHHHHHHHHH | c1a-4 | multi-selected | 0.013 | 0.022 | 0.013 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 27 | fp_O | Q9QUS1 | 251 | LIIHFHKTLKRLQLQE +++ | HHHHHHHHCCCCCCCH | c1a-5 | multi-selected | 0.011 | 0.023 | 0.042 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 28 | fp_O | Q9QUS1 | 251 | LIIHFHKTLKRLQLQE +++ | HHHHHHHHCCCCCCCH | c1d-AT-5 | multi | 0.011 | 0.023 | 0.042 | ORD | MID|NR_LBD superfamily; | 0.0 |
| 29 | fp_D | Q9QUS1 | 266 | EPEYALMAAMALFS | HHHHHHHHHHHHHC | c2-AT-4 | uniq | 0.01 | 0.018 | 0.106 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 30 | fp_D | Q9QUS1 | 287 | QREEIDQLQEEVALIL | CHHHHHHHHHHHHHHH | c1d-4 | multi | 0.033 | 0.045 | 0.26 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 31 | fp_D | Q9QUS1 | 290 | EIDQLQEEVALILN | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.029 | 0.031 | 0.203 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 32 | fp_D | Q9QUS1 | 313 | LQSRFLYAKLMGLLA | CCCCCCHHHHHHHHH | c1b-AT-4 | multi | 0.192 | 0.069 | 0.037 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 33 | fp_D | Q9QUS1 | 314 | QSRFLYAKLMGLLA | CCCCCHHHHHHHHH | c2-AT-4 | multi-selected | 0.186 | 0.06 | 0.033 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 34 | fp_D | Q9QUS1 | 321 | KLMGLLAELRSINS | HHHHHHHHHHHHHH | c3-4 | uniq | 0.164 | 0.022 | 0.025 | boundary | MID|NR_LBD superfamily; | 0.0 |
| 35 | fp_D | Q9QUS1 | 345 | GLSAMMPLLGEICS | CCCCCCHHHHHHCC | c3-4 | uniq | 0.022 | 0.11 | 0.063 | __ | boundary|NR_LBD superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment