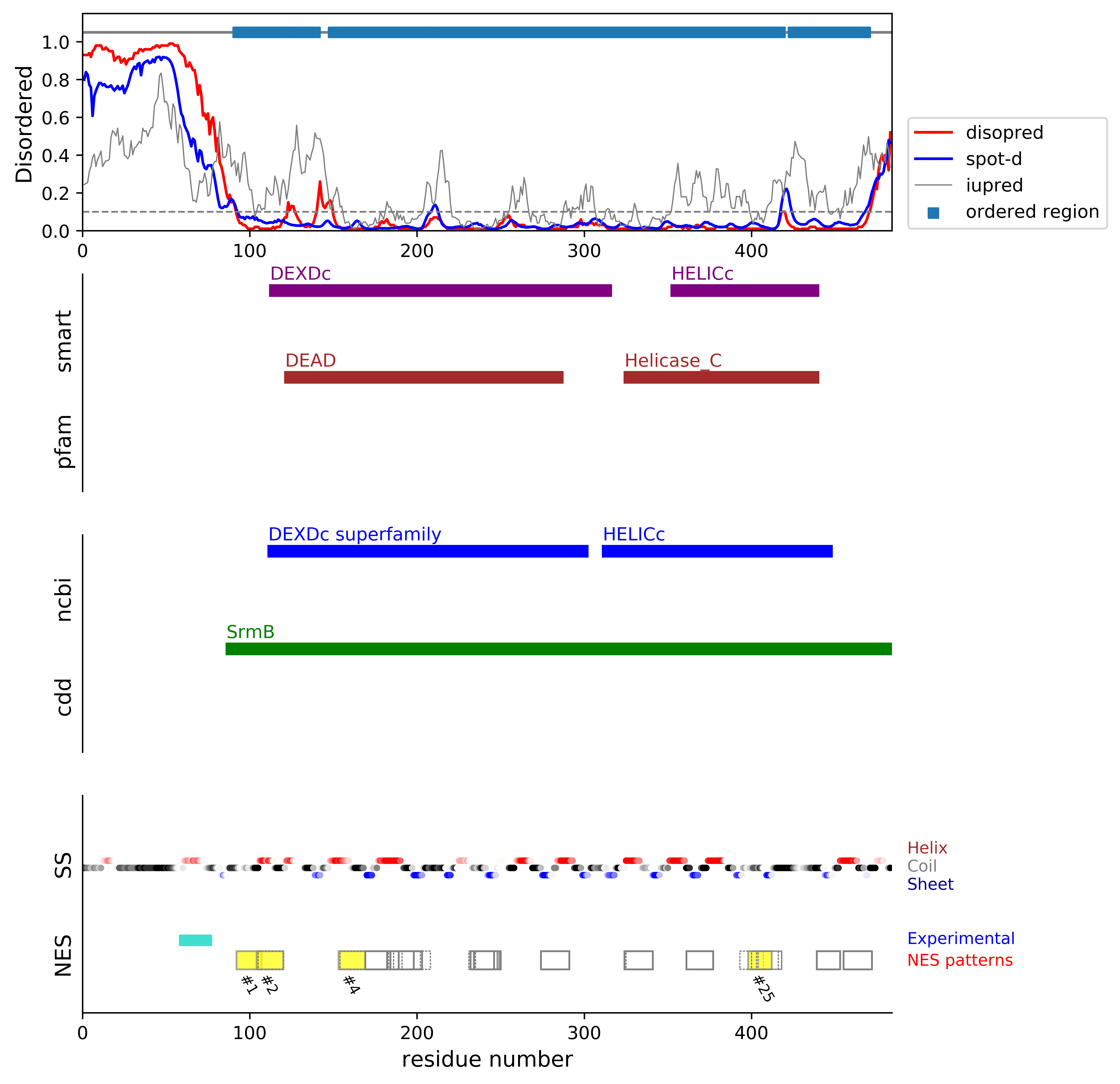

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | Q9QY16 | 92 | PLYSVKTFEELRLKE | CCCCCCCCCCCCCCH | c1b-5 | uniq | 0.028 | 0.071 | 0.244 | boundary | boundary|SrmB; | 0.0 |
| 2 | fp_D | Q9QY16 | 104 | LKEELLKGIYAMGFNR | CCHHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.016 | 0.049 | 0.179 | boundary | boundary|SrmB; | 0.0 |
| 3 | fp_D | Q9QY16 | 105 | KEELLKGIYAMGFNR | CHHHHHHHHHCCCCC | c1b-4 | multi | 0.015 | 0.048 | 0.184 | boundary | boundary|SrmB; | 0.0 |
| 4 | fp_D | Q9QY16 | 153 | FVLAMLNRVNALELFP | HHHHHHHCCCCCCCCC | c1a-5 | multi-selected | 0.011 | 0.026 | 0.056 | boundary | MID|SrmB; | 0.0 |
| 5 | fp_D | Q9QY16 | 154 | VLAMLNRVNALELFP | HHHHHHCCCCCCCCC | c1b-5 | multi | 0.011 | 0.026 | 0.047 | boundary | MID|SrmB; | 0.0 |
| 6 | fp_O | Q9QY16 | 169 | QCLCLAPTYELALQT | EEEEECCCHHHHHHH | c1b-AT-4 | uniq | 0.027 | 0.011 | 0.064 | ORD | MID|SrmB; | 0.143 |
| 7 | fp_O | Q9QY16 | 182 | QTGRVVERMGKFCVDV | HHHHHHHHHCCCCCCC | c1a-4 | multi-selected | 0.02 | 0.017 | 0.078 | ORD | MID|SrmB; | 0.0 |
| 8 | fp_O | Q9QY16 | 183 | TGRVVERMGKFCVDV | HHHHHHHHCCCCCCC | c1b-4 | multi | 0.017 | 0.017 | 0.073 | ORD | MID|SrmB; | 0.0 |
| 9 | fp_O | Q9QY16 | 186 | VVERMGKFCVDVEVMY | HHHHHCCCCCCCEEEE | c1d-5 | multi | 0.013 | 0.017 | 0.063 | ORD | MID|SrmB; | 0.0 |
| 10 | fp_O | Q9QY16 | 189 | RMGKFCVDVEVMYA | HHCCCCCCCEEEEE | c3-4 | multi-selected | 0.01 | 0.017 | 0.061 | ORD | MID|SrmB; | 0.143 |
| 11 | fp_beta_O | Q9QY16 | 191 | GKFCVDVEVMYAIRGNR | CCCCCCCEEEEEECCCC | c1cR-4 | multi | 0.014 | 0.028 | 0.095 | ORD | MID|SrmB; | 0.625 |
| 12 | fp_O | Q9QY16 | 231 | FKRKLIDLTKIRVFVLD | HHCCCCCCCCCEEEEEE | c1c-AT-4 | multi | 0.01 | 0.023 | 0.026 | ORD | MID|SrmB; | 0.125 |
| 13 | fp_O | Q9QY16 | 231 | FKRKLIDLTKIRVFV | HHCCCCCCCCCEEEE | c1b-4 | multi | 0.01 | 0.025 | 0.022 | ORD | MID|SrmB; | 0.0 |
| 14 | fp_O | Q9QY16 | 232 | KRKLIDLTKIRVFVLD | HCCCCCCCCCEEEEEE | c1d-AT-4 | multi | 0.01 | 0.023 | 0.024 | ORD | MID|SrmB; | 0.143 |
| 15 | fp_O | Q9QY16 | 232 | KRKLIDLTKIRVFV | HCCCCCCCCCEEEE | c2-4 | multi-selected | 0.01 | 0.025 | 0.02 | ORD | MID|SrmB; | 0.0 |
| 16 | fp_O | Q9QY16 | 234 | KLIDLTKIRVFVLDEA | CCCCCCCCEEEEEECC | c1aR-4 | multi-selected | 0.012 | 0.02 | 0.025 | ORD | MID|SrmB; | 0.429 |
| 17 | fp_O | Q9QY16 | 234 | KLIDLTKIRVFVLDE | CCCCCCCCEEEEEEC | c1b-5 | multi | 0.011 | 0.021 | 0.023 | ORD | MID|SrmB; | 0.429 |
| 18 | fp_O | Q9QY16 | 235 | LIDLTKIRVFVLDE | CCCCCCCEEEEEEC | c2-AT-5 | multi | 0.011 | 0.02 | 0.023 | ORD | MID|SrmB; | 0.429 |
| 19 | fp_O | Q9QY16 | 235 | LIDLTKIRVFVLDE | CCCCCCCEEEEEEC | c3-AT | multi | 0.011 | 0.02 | 0.023 | ORD | MID|SrmB; | 0.429 |
| 20 | fp_beta_O | Q9QY16 | 274 | QMLLFSATFEDSVWQFA | CEEEEEEECCHHHHHHH | c1cR-5 | uniq | 0.011 | 0.019 | 0.063 | ORD | MID|SrmB; | 0.5 |
| 21 | fp_O | Q9QY16 | 324 | DKYQALCNIYGGITIGQ | CHHHHHHHHHCCCCCCC | c1c-AT-4 | multi-selected | 0.01 | 0.017 | 0.048 | ORD | MID|SrmB; | 0.0 |
| 22 | fp_O | Q9QY16 | 325 | KYQALCNIYGGITIGQ | HHHHHHHHHCCCCCCC | c1d-4 | multi | 0.01 | 0.017 | 0.049 | ORD | MID|SrmB; | 0.0 |
| 23 | fp_beta_O | Q9QY16 | 361 | DGHQVSLLSGELTVEQ | CCCEEEEECCCCCHHH | c1d-4 | uniq | 0.017 | 0.029 | 0.198 | ORD | MID|SrmB; | 0.571 |
| 24 | fp_D | Q9QY16 | 393 | ITTNVCARGIDVKQ | EECCCCCCCCCCCC | c2-AT-4 | multi | 0.01 | 0.029 | 0.114 | boundary | MID|SrmB; | 0.0 |
| 25 | fp_D | Q9QY16 | 398 | CARGIDVKQVTIVV | CCCCCCCCCCEEEE | c2-4 | multi-selected | 0.01 | 0.028 | 0.087 | boundary | MID|SrmB; | 0.0 |
| 26 | fp_D | Q9QY16 | 400 | RGIDVKQVTIVVNFDL | CCCCCCCCEEEEECCC | c1a-AT-4 | multi | 0.011 | 0.024 | 0.119 | boundary | MID|SrmB; | 0.429 |
| 27 | fp_D | Q9QY16 | 400 | RGIDVKQVTIVVNFDL | CCCCCCCCEEEEECCC | c1d-4 | multi | 0.011 | 0.024 | 0.119 | boundary | MID|SrmB; | 0.429 |
| 28 | fp_beta_D | Q9QY16 | 403 | DVKQVTIVVNFDLPV | CCCCCEEEEECCCCC | c1b-5 | multi | 0.015 | 0.025 | 0.152 | boundary | MID|SrmB; | 0.714 |
| 29 | fp_beta_D | Q9QY16 | 404 | VKQVTIVVNFDLPV | CCCCEEEEECCCCC | c2-AT-4 | multi | 0.016 | 0.024 | 0.158 | boundary | MID|SrmB; | 0.714 |
| 30 | fp_beta_D | Q9QY16 | 439 | GKKGLAFNMIEVDK | CCCCCEEEEECCCC | c2-4 | uniq | 0.01 | 0.034 | 0.152 | boundary | MID|SrmB; | 0.714 |
| 31 | fp_D | Q9QY16 | 455 | LLMKIQDHFNSSIKQLD | HHHHHHHHHCCCCEECC | c1cR-5 | uniq | 0.028 | 0.064 | 0.284 | boundary | boundary|SrmB; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment